Figure 1.
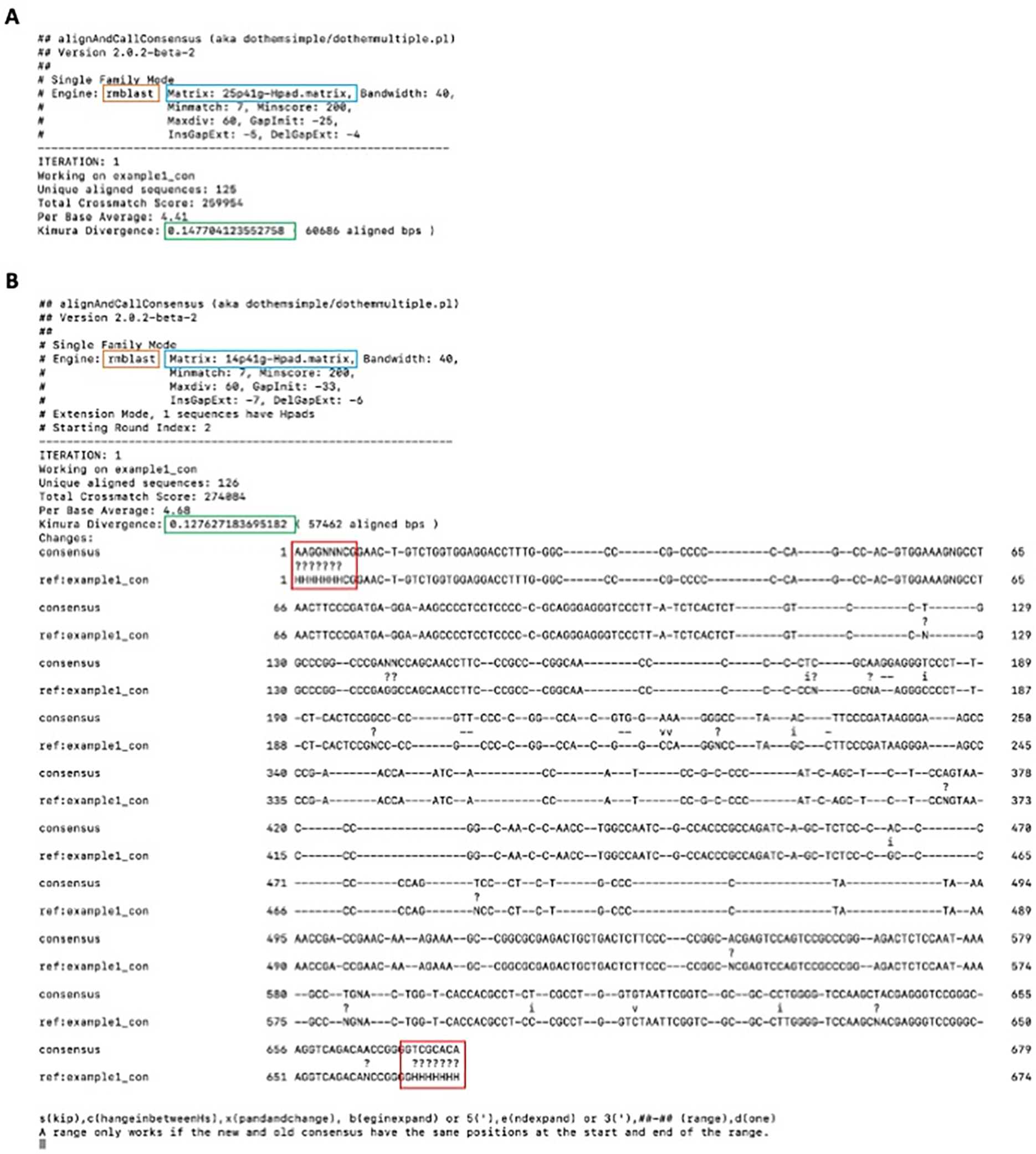
Terminal output of alignAndCallConsensus.pl performed with A) the default 25% substitution matrix and B) a 14% substitution matrix. The orange box indicates the search engine utilized, while the blue box indicates the substitution matrix used for this alignment. 25p41g indicates that a 25% substitution matrix with a 41% CG background was used. The green box highlights the average Kimura-model substitution level of all the aligned copies compared to the consensus. The presented sequences are the newly calculated consensus (“consensus”) and the previous consensus or reference sequence (“ref:example1_com”) as they appear in the complete MSA (gaps in both sequences indicate that copies exist with an insertion ther). “v” in between the sequences indicates a transversion, while an “i” indicates a transition. “?” indicates a nucleotide aligned to the ambiguous base (like “N” or “H”). The red boxes highlight the consensus of the bases aligned to the terminal “H-pads”.
