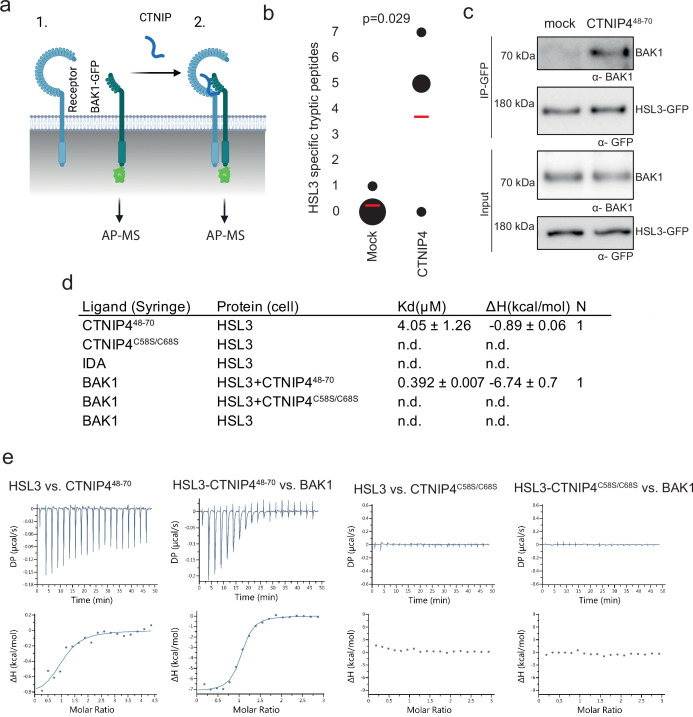
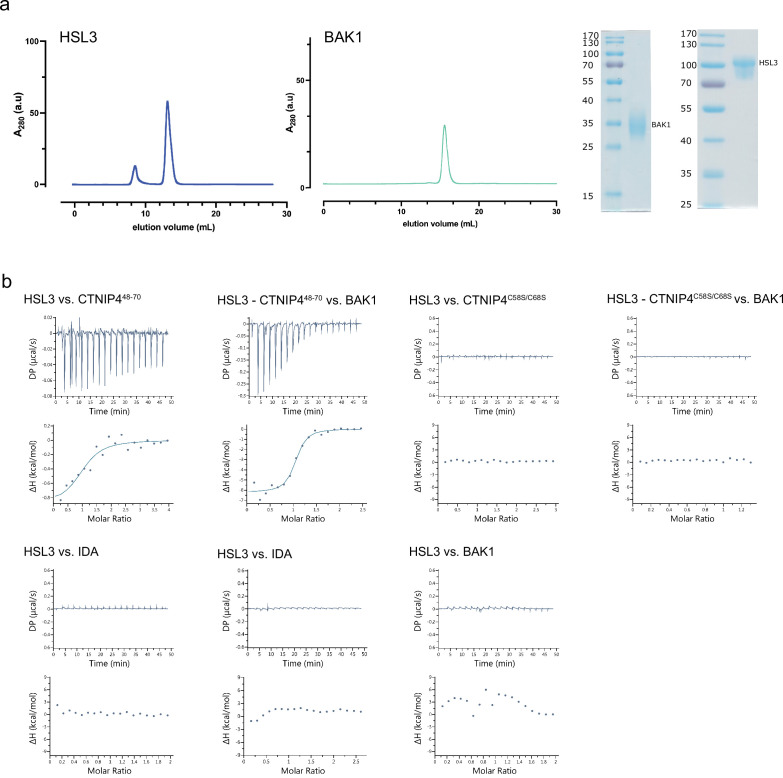
Figure 2. HAESA-LIKE 3 (HSL3) forms a CTNIP-induced receptor complex with BAK1.
(a) Schematic representation of BAK1-GFP immunoprecipitation in the (1) absence or (2) presence of CTNIP4 treatment to identify protein associations induced by CTNIP. Figure generated using Biorender. (b) HSL3-specific spectral counts identified in four independent biological replicates where BAK1-GFP was pulled down in the presence or absence of 1 μM CTNIP4 treatment. Circle diameter is proportional to the number of replicates. Red lines indicate the mean spectral counts for each treatment. p-Values indicate significance relative to the untreated control in a two-tailed t-test. (c) Affinity purification of BAK1 with HSL3-GFP from HSL3-GFP seedlings treated with 1 μM CTNIP448-70 or water for 10 min. Western blots were probed with antibodies α-GFP and α-BAK1. This experiment was repeated three times with similar results. (d) Isothermal titration calorimetry (ITC) summary table of HSL3 vs. CTNP448-70, CTNP4C58S/C68S and INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptides, and contribution of the BAK1 co-receptor to the ternary complex formation. Kd, (dissociation constant) indicates the binding affinity between the two molecules considered (nM). The N indicates the reaction stoichiometry (N=1 for a 1:1 interaction). The values indicated in the table are the mean ± SEM of two independent experiments. (e) ITC experiments of HSL3 vs. CTNIP4 and CTNIP4C58S/C68S, in the absence and presence of the co-receptor BAK1. GFP, green fluorescent protein.