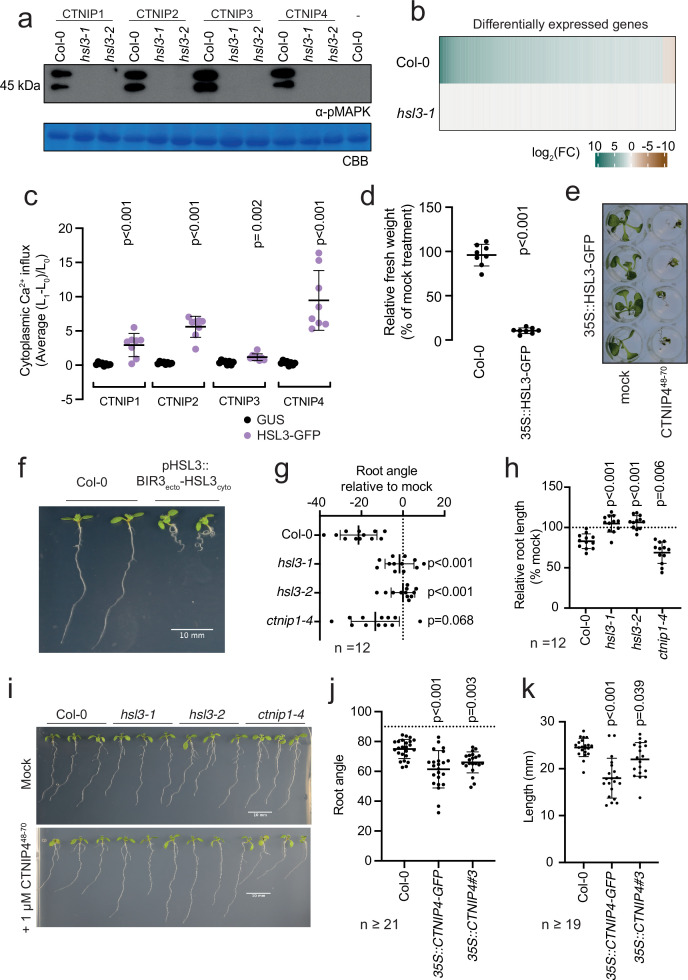
Figure 3. HAESA-LIKE 3 (HSL3) is strictly required for CTNIP perception and growth regulation.
(a) Western blot using α-p42/p44-ERK recognising phosphorylated MAP kinases in seedlings treated with 100 nM CTNIPs or mock for 15 min. The membrane was stained with Coomassie brilliant blue (CBB), as a loading control. (b) Heat map showing all significantly differentially expressed genes (p<0.05, |log2(FC)|>1) in Arabidopsis wild-type (WT) or hsl3-1 seedlings treated with or without 100 nM CTNIP448-70 for 30 min relative to a mock control. (c) Mean relative cytoplasmic Ca2+ influx in leaf disks of Nicotiana benthamiana transiently expressing the defined constructs induced by 1 μM CTNIP or mock application (n = 8 leaf disks). A line represents mean; error bars represent SD. p-Values indicate significance relative to the GUS-transformed control in a Dunnett’s multiple comparison test following one-way ANOVA. (d) Fresh weight of 14-day-old seedlings grown in the presence of 500 nM CTNIP448-70 for 10 days relative to mock (n = 8 seedlings). A line represents mean; error bars represent SD. p-Values indicate significance relative to the WT control in a two-tailed t-test. (e) Representative images of (d). (f,i) Nine-day-old vertically grown Arabidopsis seedlings on 1/2 Murashige and Skoog (MS) agar medium with 1% sucrose. Pictures were taken from the front of the plate. (g–h,j–k) Root parameters were quantified from the base of the hypocotyl to the root tip using ImageJ. (g) Root angle is shown relative to mock. Negative values indicate leftward root skewing. (j) Absolute root angle with 90° representing the gravity vector. Angles < 90° represent skewing to the left. A line represents mean; error bars represent SD. p-Values indicate significance relative to the WT control in a Dunnett’s multiple comparison test following one-way ANOVA. All experiments were repeated and analysed three times with similar results.
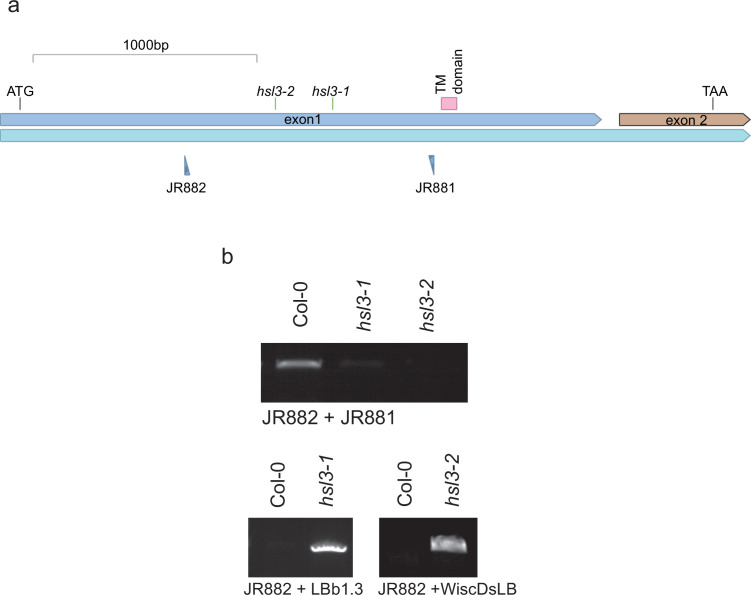
Figure 3—figure supplement 1. Genetic characterisation of hsl3 mutants.
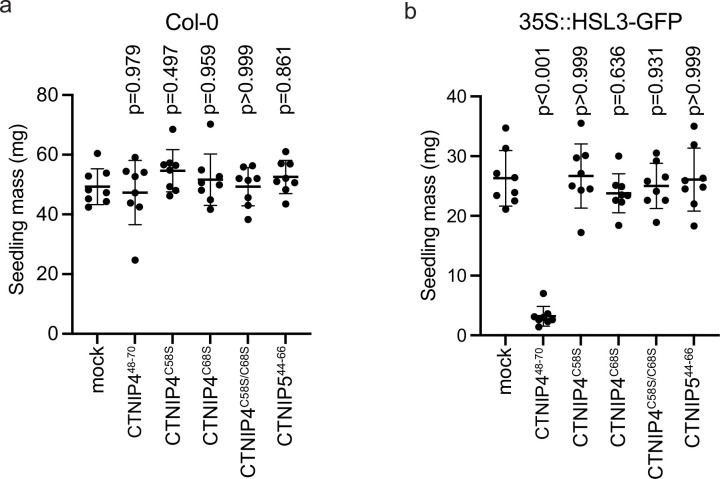
Figure 3—figure supplement 2. CTNIP-induced seedling growth inhibition.
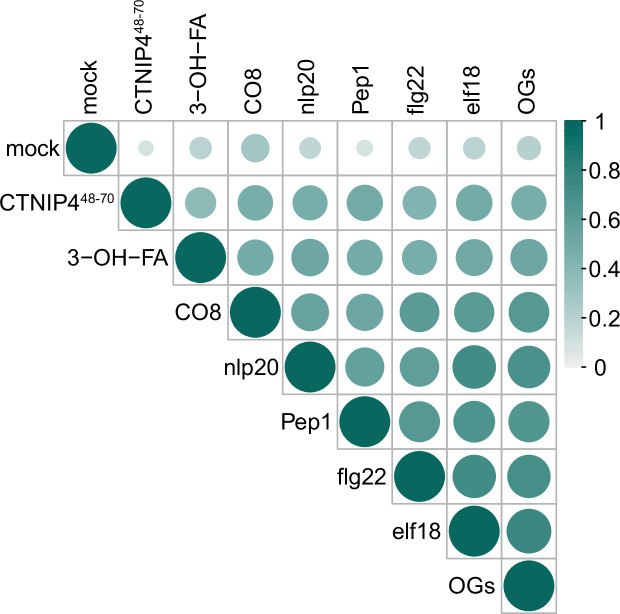
Figure 3—figure supplement 3. Correlation of CTNIP4-induced transcriptomic response with that of elicitors at 30min.
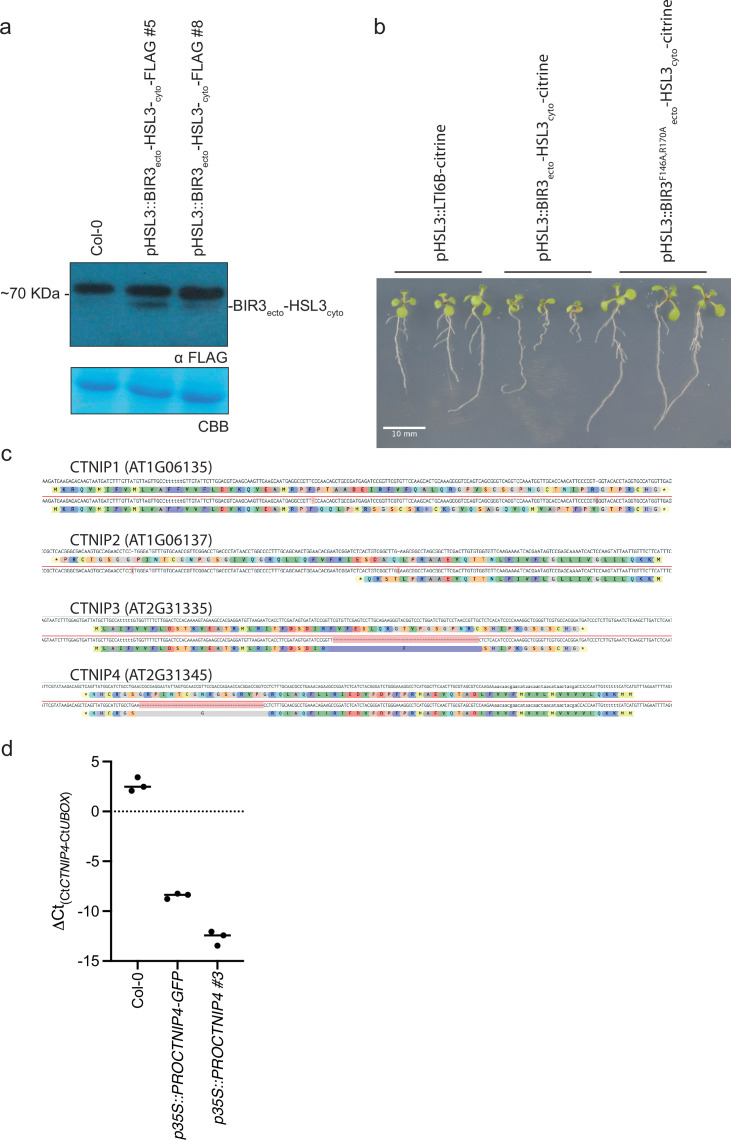
Figure 3—figure supplement 4. Characterisation of CTNIP and chimeric receptor lines.
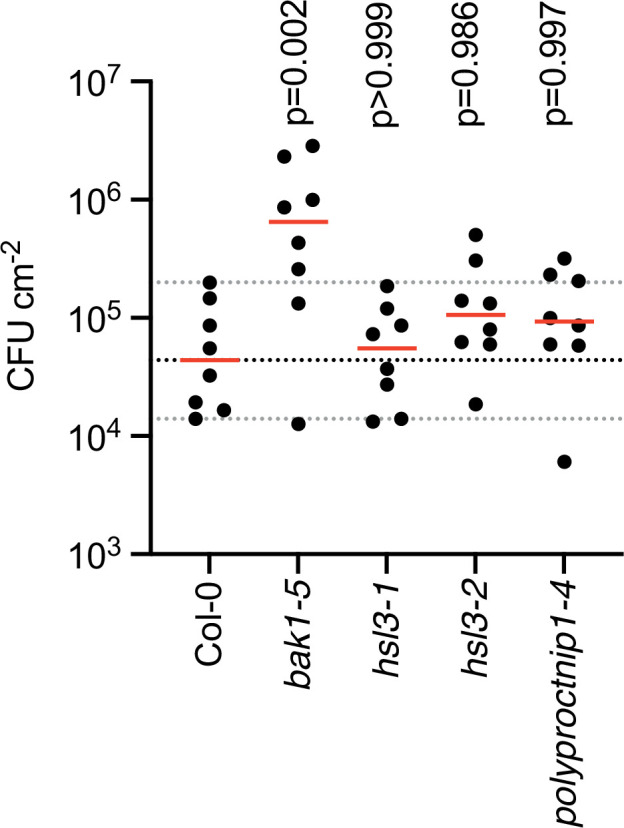
Figure 3—figure supplement 5. HSL3-CTNIP mutants do not show altered resistance to Pseudomonas syringae pv tomato DC3000 ΔAvrPto/ΔAvrPtoB.