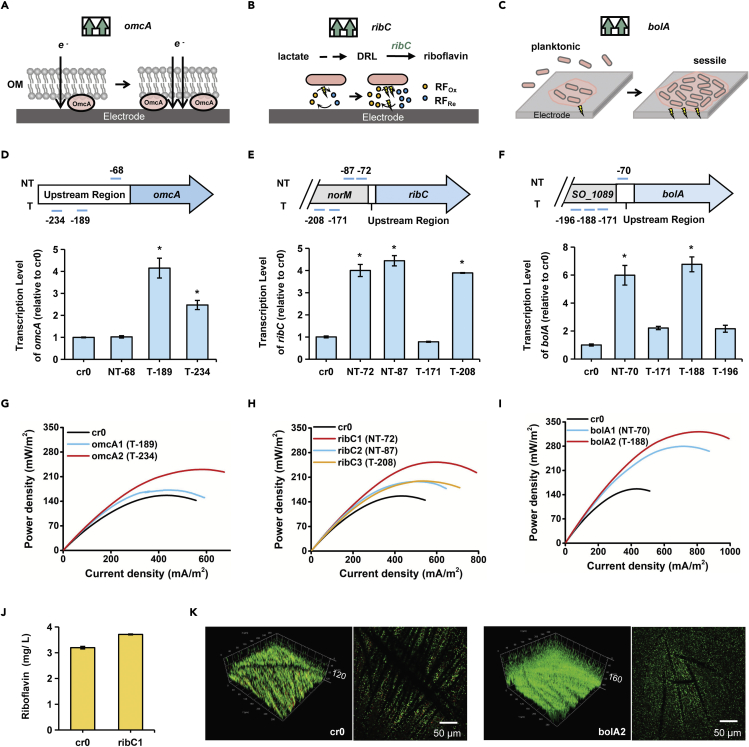
Figure 5.
Activation of cytochrome, electron shuttle, cell motility-related gene expression by PaeCascade-RpoD for enhancing EET efficiency in S. oneidensis MR-1
(A–C) Schematic representation of (A) promoting OM c-Cyts by activating the expression of omcA, (B) modulating RF biosynthesis pathway to produce more RF by activating the expression of ribC, (C) transiting engineered strains from planktonic to the attachment stage by activating the expression of bolA to enhance the EET efficiency.
(D–F) Targeting sites and corresponding transcriptional activation efficiency of (D) omcA, (E) ribC, and (F) bolA by qRT-PCR. norM and SO_1089 are the adjacent upstream genes of ribC and bolA, respectively. Transcription levels are normalized to the control strain cr0. Values and error bars indicate mean ± SEM of three replicates. Asterisks mean the strains selected for the bio-electrochemical analysis in microbial fuel cells.
(G–I) The power density output curves obtained by linear sweep voltammetry (LSV) with a scan rate of 0.1 mV/s of (G) omcA, (H) ribC, and (I) bolA-activated strains. cr0 is the control strain. See also Figure S2.
(J) The production of riboflavin (RF) of the control strain cr0 and engineered strain ribC1. Values and error bars indicate mean ± SEM of three replicates.
(K) Bio-image of strain cr0 and the activated strain bolA2 embedded on anode carbon cloth by Confocal Laser Scanning Microscope (CLSM). The number on the right of the biofilm means the bio-image thickness. All scale bars are 50 μm.