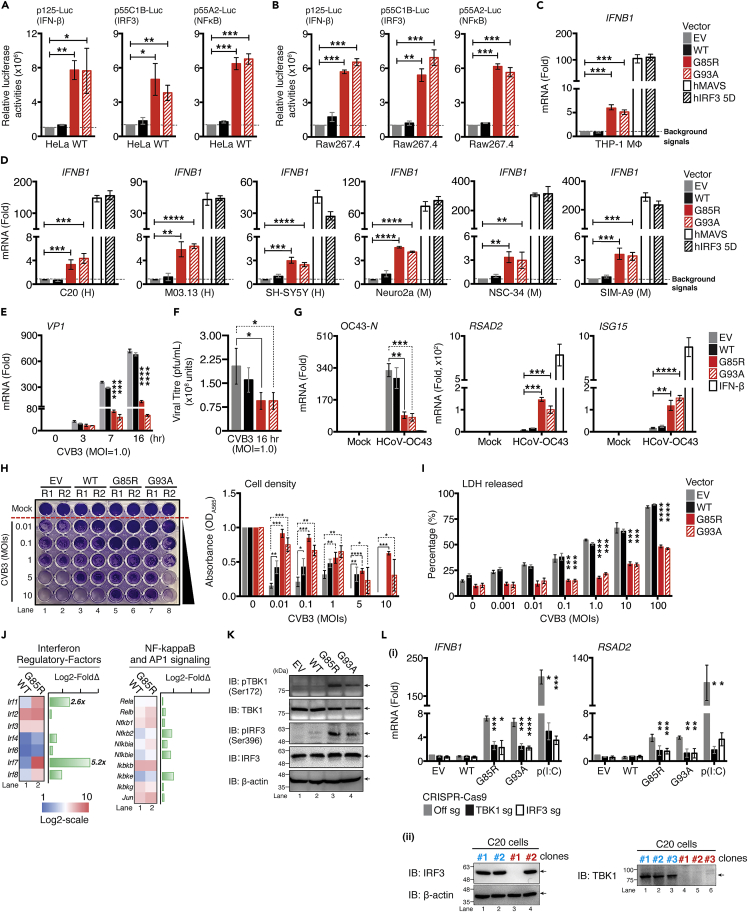
Figure 2.
Overexpression of ALS SOD1-mutants activate IFN-I response in vitro and confer resistance to viral infection
(A and B) HeLa WT or murine RAW264.7 cells expressing indicated reporter were co-transfected with plasmids encoding EGFP-, human SOD1-WT,-G85R, or-G93A mutants. 36 h after transfection, cell lysates were collected and analyzed for luciferase activity.
(C) THP-1-derived MΦ cells were transfected with 0.01 μg of indicated plasmids for 36 h, indicated mRNA levels were assessed via RT-qPCR.
(D) Similar conditions in panel (C) were performed in an indicated neuronal cell lineage (H: Human; M: Murine).
(E) C20 cells were transfected with indicated plasmids, followed by CVB3 (MOI = 1.0) infection at indicated time-points. CVB3 Vp1 mRNA was measured via RT-qPCR.
(F) Supernatant from panel E were collected and subjected to the plaque assay to measure the release of viral particles.
(G) Newly seeded C20 cells were pre-treated with supernatant collected from C20 cells expressing the indicated plasmids for 24 h. Cells were then infected with HCoV-OC43 (MOI = 1.0) for 72 h. In a parallel experiment, C20 cells were treated with 100 ng/mL of IFN-β proteins for 24 h as positive control. HCoV-OC43 nucleocapsid RNA, Isg15, and Rsad2 mRNAs were measured using RT-qPCR.
(H and I) C20 cells transfected with the indicated plasmid were infected with the indicated MOI of CVB3 and assessed 24 h later for cell viability with crystal violet staining and the LDH released assay.
(J) NanoString analysis for gene expression of indicated family of transcription factors comparing whole brain of WT and SOD1-G85R mice (n = 3 for each genotype).
(K) Immunoblot analysis of phosphorylated or total TBK1 and IRF3 or β-actin in C20 cells after transfection with the indicated plasmids.
(L) (1) TBK1, or IRF3-deficient C20 cells using CRISPR/Cas9 system expressing the indicated plasmids for 36 h. Ifnb1 and Rsad2 mRNA measured via RT-qPCR. Transfected poly(I:C) [p(I:C), 0.1 μg] served as positive control. (2) Knockout efficiency of the positive clones was confirmed by western blotting. Clone#1 of two lineage were selected for subsequent studies. Data represent the mean of three independent experiments with n = 3 biological replicates (mean ± SEM). ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001; (A–I, and L: two-tailed, unpaired Student’s t test). See also Figure S3.