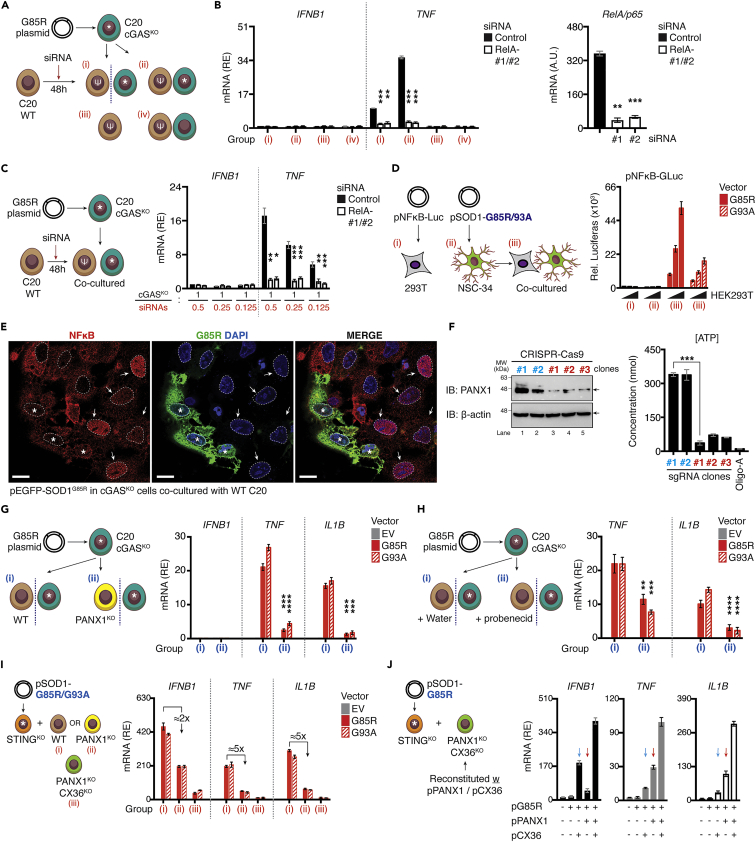
Figure 8.
Pannexin-1 and connexin-36 cooperatively propagate NF-κB dependent cytokines to bystander cells
(A and B) Schematic diagram showing the experimental design. cGAS-deficient C20 cells expressing G85R plasmid [marked with asterisk (∗)] were co-cultured with WT C20 cells transfected with 20 nM of indicated siRNAs [marked with epsilon (ψ)] in the presence or absence of the trans-well system. The expression of indicated mRNA were measured via RT-qPCR. Knockdown efficiency of RelA gene is shown (B, right).
(C) cGAS-deficient C20 cells expressing G85R plasmid [marked with asterisk (∗)] co-cultured with titrated number of WT C20 cells transfected with 20 nM of the indicated siRNAs [marked with epsilon (ψ)] in increasing ratios of (1:0.5/0.25/0.125). Expression of Ifnb1, or Tnf quantified via RT-qPCR.
(D) HEK293T cells expressing NF-κB reporter plasmid and NSC-34 cells expressing SOD1 mutants were culture alone or co-cultured before the luciferase assay.
(E) Confocal analysis of pSOD1-G85R expressing cGAS-deficient cells co-cultured with WT C20 cells. EGFP-G85R stably expressing cells are marked with asterisk (∗). Nuclear translocation of NF-κB in bystander cells is indicated by white arrows. Nuclear-membrane is marked by white-dotted lines.
(F) Knockout efficiency of PANX1 using CRISPR-Cas9 verified using immunoblotting. Amount of ATP within cultured supernatant were measured using an ATP-assay. Treatment with 1.0 μM oligomycin A served as the positive control.
(G) cGAS-deficient C20 cells expressing G85R or G93A plasmid (indicated as asterisk) were co-incubated with WT or PANX1-deficient cells in the trans-well system. Statistical analysis shown in Group (ii) was performed comparing the same condition in Group (i).
(H) Similar experimental design as in (G) was used but the cell was replaced with WT C20 cells treated with vehicle (water) or 100 μM probenecid in the trans-well system. Statistical analysis shown in Group (ii) was performed comparing the same condition in Group (i).
(I) WT, PANX1-deficient (yellow), or PANX1-CX36-double deficient (light green) C20 cells were co-cultured with STING-deficient C20 cells expressing SOD1-mutants [marked by asterisk (∗), orange]. Relative induction of the indicated mRNA from designated groups (i, ii, or iii) were quantified via RT-qPCR. Fold-reduction is indicated.
(J) PANX1-CX36-double deficient (light green) C20 cells were reconstituted with WT PANX1 or CX36 plasmids before co-culturing with STING-deficient C20 cells expressing G85R for 36 h. Relative induction of Ifnb1, Tnf, or Il1b mRNA was measured. All RT-qPCR data were normalized to β-actin. Data represent the mean of at least three independent experiments (mean ± SEM). ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001; (B, C, and F–H: unpaired Student’s t test).