Figure 5. pUb binds to the parkin RING0 domain.
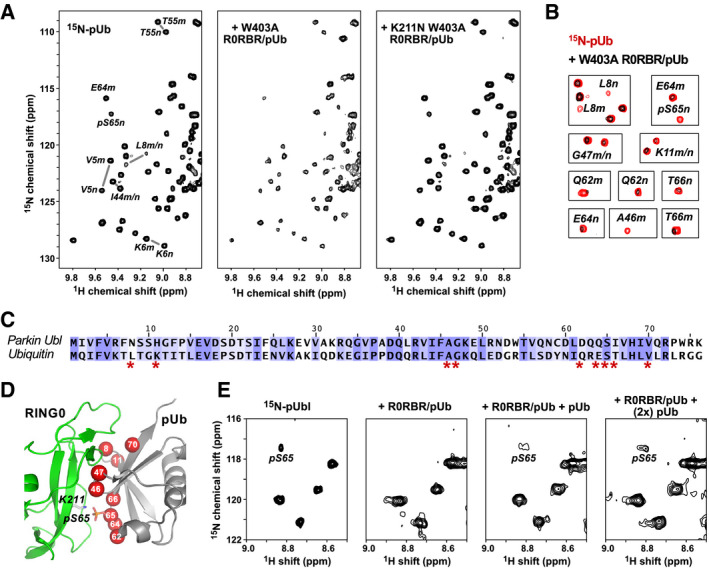
- 1H‐15N correlation spectra at 25°C of 50 µM 15N‐pUb alone (left) and in the presence of 100 µM complexes of pUb with W403A R0RBR (middle) or K211N W403A R0RBR parkin (right). Selected backbone amide signals are labeled according to residue number and the pUb conformation, major (m) or minor (n).
- Identification of 15N‐pUb signals that weaken or disappear when pUb is bound to W403A R0RBR.
- Sequence alignment of ubiquitin and the Ubl domain of human parkin colored according to similarity. Ubiquitin residues whose NMR signals change upon binding RING0 are marked by asterisks.
- Mapping of 15N‐pUb signals that weaken or disappear (red spheres) onto a model of pUb bound to parkin RING0.
- Competitive binding of pUbl and pUb to RING0. Portion of 1H‐15N NMR correlation spectra at 5°C of 60 µM 15N‐pUbl alone (left), in the presence of a 1:1 complex of rat parkin R0RBR and pUb (middle left), and in the presence of R0RBR/pUb supplemented with 60 μM pUb (middle right) and 120 μM pUb (right). The temperature was lower for this experiment to enhance detection of the weak binding by pUbl.
