Summary
In humans, one of the X chromosomes in genetic females is inactivated by a process called X chromosome inactivation (XCI). Variation in XCI across the placenta may contribute to observed sex differences and variability in pregnancy outcomes. However, XCI has predominantly been studied in human adult tissues. Here, we sequenced and analyzed DNA and RNA from two locations from 30 full-term pregnancies. Implementing an allele-specific approach to examine XCI, we report evidence that XCI in the human placenta is patchy, with large patches of either maternal or paternal X chromosomes inactivated. Further, using similar measurements, we show that this is in contrast to adult tissues, which generally exhibit mosaic X inactivation, where bulk samples exhibit both maternal and paternal X chromosome expression. Further, by comparing skewed samples in placenta and adult tissues, we identify genes that are uniquely inactivated or expressed in the placenta compared with adult tissues, highlighting the need for tissue-specific maps of XCI.
We confirm and extend previous findings, presenting evidence for patchiness of X inactivation in the human placenta. We show that this is unique to the placenta compared with adult tissues but that most genes, with five notable exceptions, show similar escape or inactive status in the placenta and adult tissues. Keywords: placenta; X-inactivation, somatic; escape; X chromosome
Introduction
X chromosome inactivation (XCI) evolved in therian mammals at the same time as the development of invasive placentation and is important for many biological processes.1 XCI is a mechanism to regulate the dosage of gene expression due to the differences in the number of X chromosomes between the sexes: genetic females (XX) have two X chromosomes while genetic males (XY) have only one X chromosome.2,3
There are two levels of XCI. The first level is at the whole-chromosome level, where either the maternal X or the paternal X is chosen for inactivation. The choice of which X is inactivated has been observed to happen in one of two ways: random XCI (the maternal and paternal X are inactivated with equal probability) and imprinted XCI (the paternal X is inactivated). In mice, the extraembryonic lineages that ultimately give rise to the placenta and some extraembryonic membranes show paternally imprinted XCI.4, 5, 6 Paternally imprinted XCI has also been reported in rats,7 cows,8 and marsupial mammals.9, 10, 11 However, in mice, the embryonic lineages that ultimately give rise to the rest of the fetus exhibit random XCI.4 Random XCI has also been reported in mule and horse placenta.12
The second level of XCI concerns which individual genes are subject to inactivation (silenced or not expressed) on the inactive X chromosome. Previous studies have aimed to determine what genes on the X chromosome escape XCI in a variety of tissues (see Carrel and Brown13 for an overview of expression from the inactive X). Carrel and Willard14 studied biallelic expression in primary human fibroblast cell lines and rodent and human somatic hybrids. Cotton et al.15 used DNA methylation data to characterize escape status in 27 adult tissues. Gene-specific inactivation has also been shown to vary between tissues and individuals in mouse and human.16,17 However, previous cross-tissue studies on gene-specific escape from XCI did not include the placenta.
The human placenta is a transient tissue that is formed early in pregnancy and develops from the outer layer of the pre-implantation embryos.18,19 The placenta has the genotype of the fetus and plays a critical role in pregnancy by regulating nutrition and protecting the developing fetus from the pregnant person’s immune system.20 Improper placenta development can lead to complications, such as pre-eclampsia and fetal growth restriction.21 It is important to study gene-specific escape in the placenta because XCI could play an important role in pregnancy complications. Gong et al.22 found that the spermine synthase gene (SMS) that modulates fetal growth restriction, a common pregnancy complication, may escape XCI in the placenta, but not in other tissues. Further, the degree of XCI skewing could be associated with pregnancy loss.23 Therefore, understanding XCI in this early formed tissue is critical for understanding downstream developmental effects.
Previous research suggests that XCI across the entire X chromosome in the placenta is random and patchy, but these studies relied on a limited number of loci and did not investigate chromosome-wide, gene-specific variability or compare with adult tissues.24,25 Further, one of the challenges of previous research on XCI is that indirect methods have been used to infer inactivation. Gong et al.22 and Tukiainen et al.17 used higher expression in females as compared with males as a proxy for escape genes, rather than directly measuring inactivation status. Slavney et al.26 observed that escape genes tend to show higher expression compared with inactivated genes in both males and females but with little ability to discriminate between individual gene escape and inactivation, especially for heterogeneously escaping genes.
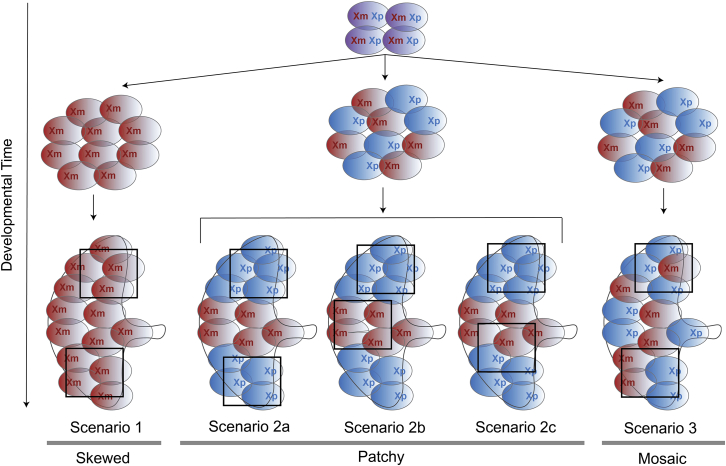
Here, we utilized next-generation sequencing to characterize genome-wide patterns of XCI in the human placenta. We further compared patterns of XCI between the placenta and adult tissues in humans and analyzed genes with an XCI status that is unique to the placenta. We studied allele-specific expression from 30 full-term placentas from uncomplicated pregnancies by performing whole-exome and whole-transcriptome (from two locations) sequencing, where the sex of the offspring was assigned female at birth. We further utilized data from the Genotype-Tissue Expression (GTEx) project to study and compare patterns of XCI in adult tissues.27 Using our experimental approach, we have the capacity to observe (1) extreme skewing in the human placenta, where all placentas show the same X inactivated in both sites; (2) patchiness, where the placenta exhibits patches of daughter cells either maternal or paternal X expression only; or (3) mosaicism, where clusters of cells in a sample exhibit both maternal and paternal X chromosome expression (Figure 1). In this study, we limited analysis of gene-specific inactivation to placental and adult tissues with skewed XCI to directly measure allele-specific expression. We observed evidence for skewing (large patches of exclusively maternal or paternal X expression) in the human placenta that is not present in adult tissues. We also compared genes that are inactivated, genes that escape from inactivation, and genes that show variable escape between the placenta and adult tissues. While a majority of genes are concordant for inactivation status between the placenta and adult tissues (71%), we observed a subset of genes that show opposite patterns between the placenta and adult tissues.
Figure 1.
Schematic possible patterns of X chromosome inactivation
Prior to XCI, both the maternal and the paternal X chromosomes are present (purple cells). The blue cell represents the maternal X is inactivated, and the red cell represents the paternal X is inactivated. If XCI occurs very early in development and propagates to all daughter cells, we would observe extreme skewing, where all placentas show the same X inactivated. In this case, the same X chromosome is inactivated in both sites sampled from this study. If XCI occurs at an intermediate stage, XCI is random but is present with large patches of daughter cells with only the maternal X or paternal X. In this case, three possible scenarios can happen: (1) both of the sites sampled in this study happen to come from two patches with the same X inactivated, (2) each site sampled in this study comes from two patches with different X inactivation, and (3) one of the sites sampled in this study comes from the boundary of two patches. If XCI occurs very late in development, there are either small or no patches, creating a mosaic pattern of XCI.
Our results provide additional evidence that XCI in the human placenta is random and patchy. Our study further shows that patterns of X inactivation differ between embryonic tissue and adult tissues, consistent with different developmental structures of these tissues. Finally, we identified genes with unique XCI status in the placenta. We hypothesize that these genes could play an important role in the placenta and pregnancy.
Material and methods
Samples collection
Working with the Yale Biobank, we collected samples from 30 placentas, where the sex of the offspring was assigned female at birth. We confirmed that the samples were genetically XX by the presence of heterozygous sites across the X chromosome. The placenta samples were collected immediately following birth from term (≥36 weeks and 6 days) uncomplicated pregnancies delivered by cesarean section. Placentas were collected and sequenced at two different times, with 12 placenta samples in the first batch (OBG0044, OBG0068, OBG0111, OBG0115, OBG0120, OBG0133, OBG0156, OBG0170, OBG0174, OBG0175, OBG0178, and OBG0166) and 18 in the second (OBG0022, OBG0024, OBG0026, OBG0028, OBG0030, OBG0039, OBG0050, OBG0051, OBG0066, OBG0121, OBG0138, OBG0180, OBG0188, OBG0201, OBG0205, OBG0289, OBG0338, and OBG0342).
Placental tissue samples were obtained through rapid sampling (≤30 min post-delivery). Tissue samples were taken from the “maternal” side, midway between the chorionic and basal plates, from the periphery of the lobules, avoiding maternal tissue, consistent with best practices.28 Sampling sites were free of visible infarction, calcification, hematoma, and tears (areas of frank visible pathology were avoided). Whenever possible, tissue samples were obtained from distinct cotyledons of the placenta in opposing quadrants of the placenta (far from one another spatially).
The sampling protocol was as follows:
-
1.
Orient the placenta maternal side up (basal plate uppermost) and identify sampling areas in each of the four placental quadrants.
-
2.
At each site, remove the basal plate (to remove maternal tissue) ∼1 to 2 mm by trimming with a pair of sterile scissors to expose villous tissue.
-
3.
Cut a “grape-sized” (approximately 1 to 2 cm3; 5 to 6 g) tissue lobule from each of the four quadrants.
-
4.
Wash the tissue thoroughly twice in phosphate-buffered saline (PBS) solution and blot on clean gauze.
-
5.
From each grape-sized lobule, cut away eight smaller pieces (∼1 to 2 mm3) using a scalpel.
-
6.
For each sampling quadrant, place four of the ∼50-mg tissue pieces in a labeled cryovial. Snap freeze immediately in liquid nitrogen. Store samples at −80°C until ready for use; aliquot individual tissue pieces as needed per research protocols.
-
7.
For each sampling quadrant, place four of the tissue pieces in a labeled cryovial containing 1 mL of RNAlater solution (RNA-stabilizing agent). Store the cryovials in the 4°C benchtop fridge for a minimum of 48 h, and a maximum of 7 days, per RNAlater manufacturer protocol. After a minimum of 48 h, use a pipette to remove the RNAlater solution from the cryovials and immediately snap freeze in liquid nitrogen. Store samples at −80°C until ready for use; aliquot individual tissue pieces as needed per research protocols.
Whole-exome sequencing
DNA was extracted from one flash-frozen collection site for each individual. Exome libraries were prepped and sequenced to approximately 50× coverage with 100-bp paired-end sequence on the Illumina NextSeq at the Yale Genome Sequence Center.
Whole-transcriptome sequencing
From each placenta, two separate sites from opposite quadrants were collected. RNA was extracted, and RiboZero stranded libraries (RF) were prepared and sequenced to approximately 40 million reads per sample with 100-bp paired-end sequence on the Illumina NovaSeq at the Yale Genome Sequence Center.
Exome sequence data processing
We used fastqc v.0.11.829 for quality control and aggregated results from fastqc by using multiqc v.0.9.30 We trimmed adapters using bbduk as part of bbmap v.38.22.31 Sequences were trimmed both left and right for phred quality of 30, minimum length of 75 bp, and average read quality of 20 or greater to keep only high-quality reads.31 Quality was checked after trimming (Figure S1). We confirmed genetic XX females by examining the reads mapped ratio between the X chromosome and chromosome 19 (chrX/chr19), between the Y chromosome and chromosome 19 (chrY/chr19), and between the Y chromosome and the X chromosome (chrX/chrY). We observed that sample OBG0175 has a lower chrX/chr19 reads mapped ratio than other samples in this study (Figure S2). Therefore, we removed sample OBG0175 from further analyses. We used bwa-mem v.0.7.1732 to map to a female-specific reference genome. Specifically, we mapped the exome samples to a sex chromosome complement informed reference genome in which the Y chromosome is hard masked (to avoid mismapping of X-linked reads to homologous regions on the Y chromosome in the XX samples). Mapping exome samples to a reference genome with the Y chromosome hard masked has been shown to increase the number of variants genotyped on the X chromosome.33 To generate the sex chromosome complement reference genome, we employed XYalign.33 XYalign created a Y-masked gencode GRCh38.p12 human reference genome for aligning XX individuals.34 We used picard v.2.18.2735 to mark PCR duplicates. To genotype variants, we used GATK v.4.1.0.0.36, 37, 38 We first used GATK’s HaplotypeCaller to generate GVCF files. Second, we combined GVCF from 66 samples using GATK’s CombineGVCFs (30 placenta samples and an additional 36 samples from a separate study in the group [unpublished data] here to increase power to genotype variants). Finally, we used GATK’s GenotypeGVCFs to call variants. Following GATK’s best practice, we obtained high-quality variants by filtering using GATK’s Variant Quality Score Recalibration (VQSR). We tabulated the number of heterozygous variants for each sample in Table S1. Because we sequenced the placenta samples in two separate batches, we plotted the principal component for the exome data using the package SNPRelate in R39 and observed no separation by batch from the exome data (Figure S3).
Whole-transcriptome data processing
Samples were checked for quality using fastqc v.0.11.8,29 and results were aggregated across samples using multiqc v.0.9.30 Adapters were removed and sequences were trimmed both left and right for phred quality of 30, minimum length of 75 bp, and average read quality of 20 or greater using bbduck v.38.22.31 Quality was checked after trimming (Figure S4). Trimmed RNA sequencing (RNA-seq) female placenta reads were then aligned to a sex chromosome complement informed reference genome with the Y chromosome hard masked with Ns33,40 using the gencode GRCh38.p12 reference genome.34 RNA-seq samples were aligned using HISAT2,41 which has been shown to be a robust aligner for high-throughput RNA-seq reads. Total reads mapped and duplicate reads were visually checked using BAMtools stats (Table S2).42
Obtaining allele counts for allele-specific expression analysis
We used GATK ASEReadCounter (v.4.1.0.0) to obtain allele counts36 with the following thresholds: min-depth-of-non-filtered-base = 1, min-mapping-quality = 10, and min-base-quality = 10. GATK ASEReadCounter automatically filters out duplicated reads. Duplicated reads are suggested to be removed for allele-specific expression analysis.43 The number of heterozygous and expressed variants for each sample are reported in Table S3.
GTEx data processing
We downloaded ASEReadCounter counts for the X chromosome and chromosome 8 from the GTEx project, approved for project no. 8834 for General Research Use in GTEx to M.A.W. We only considered tissues with more than 10 samples per tissue and excluded two non-primary tissues: Epstein-Barr virus (EBV)-transformed lymphocytes and cultured fibroblasts, leaving 45 tissues to be analyzed in this study. We list the GTEx tissues, the number of samples per tissue examined in this study, and the number of skewed samples per tissue in Table S4. Notably, the tissue size sampled for each tissue in the GTEx dataset was approximately equal to that of the placental tissue samples collected in this study, with GTEx requiring samples to be less than 4 mm in thickness. Length may have been longer, ranging from 5 to 10 mm in length and width, though it is difficult to discern from the Tissue Harvesting Work Instruction (https://gtexportal.org/home/samplingSitePage) whether this full aliquot was used for the sequencing experiments.
Computing unphased median allele balance
For each heterozygous and expressed variant, ASEReadCounter tabulates the number of reads (the count) for the reference allele and the alternate allele. The total read count is the sum of the read count of the reference allele and the read count of the alternate allele. We define the biased allele to be the allele with more read counts. For each heterozygous and expressed variant, the unphased allele balance is the ratio between the read count of the biased allele and the total read count. Then, for each sample, the median allele balance is calculated by computing the median unphased allele balance across all heterozygous and expressed variants.
Defining threshold for allele-specific expression
To determine the threshold for defining biased allele expression, we plotted the unphased allele balance across all variants for each individual for chromosome 8 and the X chromosome (Figure S5). We observed that the unphased allele balance of most variants on the X chromosome is greater than 0.8, while the unphased allele balance of most variants on chromosome 8 is less than 0.8 (Figure S5). Therefore, in this study, we used 0.8 as a threshold for biased allele-specific expression.
Determining which X chromosome is inactivated
At the whole X chromosome level, to determine whether the same X chromosome is inactivated at the two extraction sites for each placenta sample, we employed a phasing strategy on the X chromosome by defining that the biased alleles (the alleles with higher counts) are on the same haplotype. We restricted this analysis to contain only heterozygous and expressed variants that are shared between the two extraction sites. For consistency, we defined extraction site A to be the site with more expressed variants where the unphased allele balance is greater than 0.8. We defined the activated X chromosome to consist of alleles where its allele balance (the ratio between this allele’s read count and the total read count) is greater than 0.8. In cases where the allele balance is less than 0.8, we picked an allele at random with equal probability between the reference allele and the alternate allele. Then, we computed the allele balance (defined as the ratio between the biased allele’s count and total count) for the alleles on the same X chromosome. We called this phased allele balance. At each site, we computed the median phased allele balance to use as summary statistics. To compute phased allele balance, we followed this procedure:
-
1.
Find expressed variants that are shared between site A and site B.
-
2.
Using the shared expressed variants between site A and site B, tabulate the number of expressed variants that exhibit biased expression (i.e., unphased allele balance is greater than 0.8).
-
3.
Pick a site to base phasing from based on the number of expressed variants with biased expression. For example, if site A has 50 expressed variants with biased expression and site B has 60 such variants, phasing is based on site B.
-
4.
Phasing strategy (using site B to base phasing): the expressed haplotype is generated by the following calculation: For each expressed variant that is shared between site A and site B, pick the allele with allele balance greater than 0.8 to be on the expressed haplotype. If allele balance is less than 0.8, choose an allele at random with equal probability.
Validating method based on allele-specific expression to quantify XCI
We validated our approach to determine skewness by examining the non-pseudoautosomal regions (nonPARs) of the X chromosome in males. We collected samples from 12 placentas, where the sex of the offspring was assigned male at birth. Even though the X chromosome in males is haploid, some variants are incorrectly genotyped as being heterozygous when diploidy is assumed when genotyping with GATK (Table S5). Regardless, these heterozygous variants exhibit mostly skewed expression, indicating that our method of combining both the whole-exome and whole-transcriptome sequence data is robust to determine skewness (Figure S6).
Classifying genes into genes that are inactivated, genes that escape XCI, and genes that show variable escape
At the per-gene level, we only considered samples that show skewed expression (allele balance is greater than 0.8). There are 52/58 samples in the placenta dataset and 525/4,958 samples in the GTEx dataset exhibiting skewed expression that were used for this analysis. For each gene on the X chromosome, we only considered a gene if there were at least five informative samples where in each sample there is at least one heterozygous and expressed variant for that gene. If there are more than one heterozygous and expressed variant for a gene, we summed up the counts for the biased allele and calculated allele balance by dividing the summed counts of the biased allele by the summed counts of the total counts. We categorized a gene as being inactivated if its allele balance is greater or equal to 0.8 in at least 70% of the samples and the median allele balance across all samples is greater or equal to 0.8. We categorized a gene as escaping from XCI if at most 30% of the samples have allele balance for this gene ≥0.8 and the median allele balance across all samples is ≤0.75. Otherwise, the gene is annotated as showing variable escape across all individuals. See Note S1 for more information on gene categorization and Table S6 for status of specific genes.
Coordinates used of PARs and XIST
As defined for GRCh38.p12,44 we used the following coordinates for the pseudoautosomal region 1 (PAR1), PAR2, and XIST:
PAR1: position 10,001 to position 2,781,479 on the X chromosome
PAR2: position 155,701,383 to position 156,030,895 on the X chromosome
XIST: position 73,820,651 to position 73,852,723 on the X chromosome
Results
Whole-chromosome level of X chromosome inactivation differs between the placenta and adult tissues
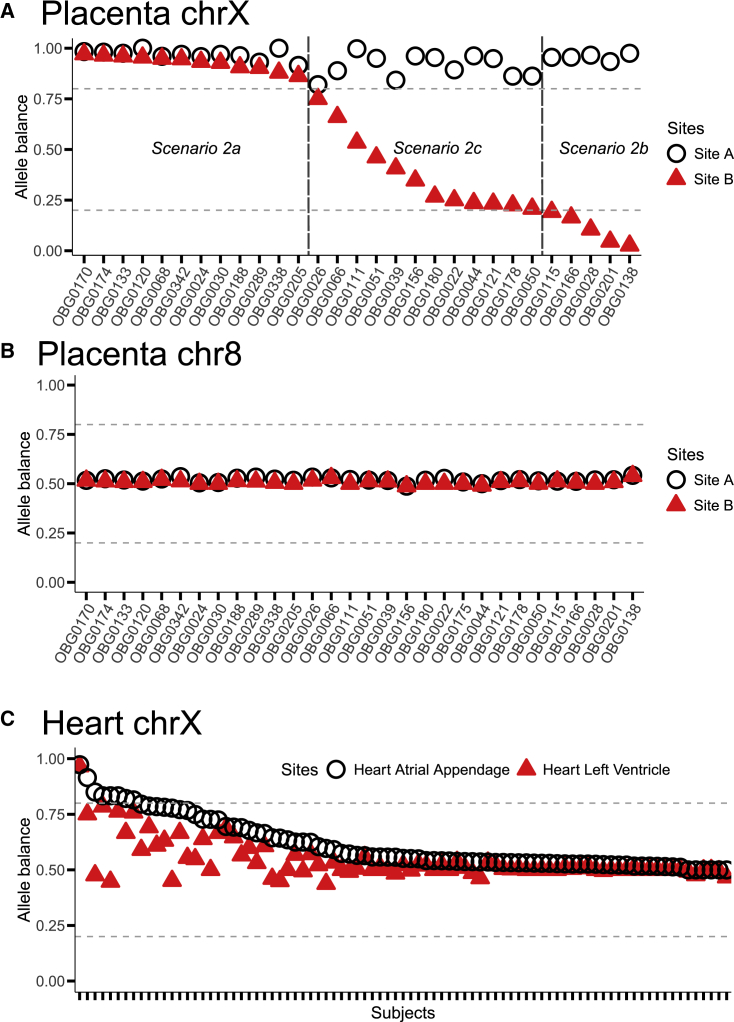
We observed evidence for patchiness in XCI in the placenta. To examine patterns of XCI in the human placenta, we sequenced the whole transcriptome from two separate extraction sites of the same placenta. We then employed a phasing strategy on the X chromosome using whole-exome and whole-transcriptome sequence data. Briefly, at heterozygous sites that exhibit allelic imbalance, we reasoned that the biased alleles (the alleles that are expressed in a higher proportion) are all on the same X chromosome (see material and methods). In 17/29 placentas, both sites in the placenta exhibited extremely skewed X inactivation, either with both extraction sites showing the same inactivated X chromosome (12 placentas; Figures 2A and S7A) or each site showing the opposite X chromosome inactivated (five placentas; Figures 2A and S7B). In the remaining 12 placentas, we observed one extraction site showing skewed XCI and the other showing variable proportions of both X chromosomes being expressed (Figures 2A and S7C). In this subset, we postulated that one of our samples was collected on the boundary of two different patches of X inactivated cells. To validate that the patterns we observed on the X chromosome are indeed XCI, we repeated the same analyses on chromosome 8, where all samples showed biallelic expression (Figure 2B). Our results suggest that the human placenta is organized into large patches with either the maternal or paternal X chromosome being inactivated.
Figure 2.
X chromosome inactivation is patchy in the human placenta and mosaic in the human heart
Phased allele balance aggregated across variants for each sample is shown for site A (open black circle) and site B (filled red triangle). Each point is the median value across variants. Median was used to minimize the effect of outliers. The dotted gray horizontal lines denote allele balance of 0.2 and 0.8.
(A) Placenta X chromosome. We removed variants that fall within the pseudoautosomal regions (PARs) prior to computing medians.
(B) Placenta chromosome 8.
(C) Heart X chromosome.
We found that patterns XCI in the placenta are different from adult tissues (Figures 2 and 3). We first compared the pattern of whole-chromosome XCI in the two placenta samples we collected with whole-chromosome XCI in two regions collected from adult human hearts (left ventricle and atrial appendages) sampled by the GTEx consortium (Figures 2A and 2C).27 We identified 85 individuals with RNA-seq data for both of these locations in the heart and employed the same phasing approach as in the placenta. In the heart tissue, both regions exhibit biallelic expression (both maternal and paternal X expression) in 91% of individuals (Figure 2C). This is in stark contrast to the placenta, where none of individuals exhibit biallelic expression at both regions (Figure 2A).
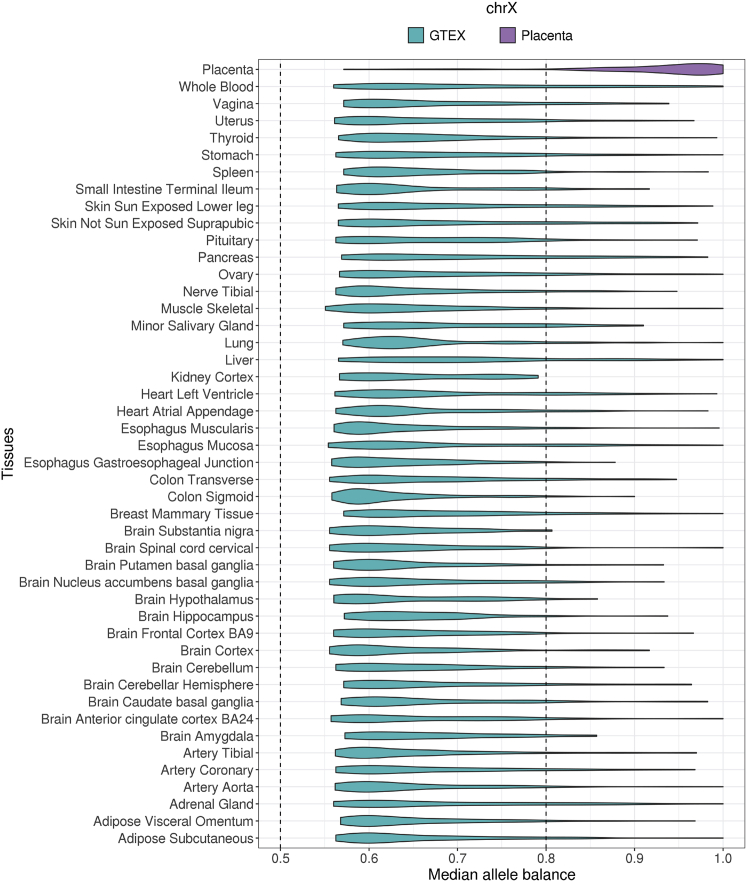
Figure 3.
Whole chromosome level of X chromosome inactivation differs between the placenta and adult tissues in humans
Unphased median allele balance is plotted for the placenta in this study (purple) and for 45 adult tissues in the GTEx dataset (blue). Each point of the violin plot is the median allele balance for each sample. Because there are no multiple site samplings for the GTEx data, unphased allele balance was computed. Here, the pseudoautosomal regions of the X chromosome are removed.
We further found the same biallelic pattern of XCI across all 43 adult tissue samples, in contrast to the placenta (Figure 3). Because other adult tissues include a single sample,27 to assess chromosome-wide XCI, we compared median allele balance computed from the biased allele for each sample. While most placenta samples (90%) exhibit skewed allele balance (i.e., allele balance greater than 0.8), most samples from adult tissues (89%) exhibit biallelic expression (i.e., allele balance between 0.5 and 0.8) (Figure 3). Similar to the placenta, however, also we observed biallelic expression across all adult tissues on chromosome 8, as expected (Figure S8). Together, these results suggest that patterns of whole-chromosome XCI differ between the placenta and adult tissues.
Gene-specific escape comparison between the placenta and adult tissues
We observed a large degree of variability in gene-specific escape in the placenta, both across and within individuals. We categorized genes on the X chromosome using only samples that exhibit skewed median allele balance across the X chromosome (i.e., median allele balance greater than 0.8; see material and methods). Only seven genes (4%) show evidence for escape across all samples with the ability to assay, while 73 genes (37%) show evidence of inactivation across all samples (Figure S9). Rather, most genes, 116 genes (59%), show a gradient of the proportion of samples that exhibit evidence for escape. In addition, heterogeneity exists between two sites within individuals (Figure S10). Given the tremendous heterogeneity in evidence for escape, we additionally investigated the ratio of female-to-male gene expression across X-linked genes. We observed that genes in all three classes (escape, variable, and inactivated) exhibit a range of female-to-male gene expression ratios, with higher gene expression in females as compared with males underpowered to determine whether a gene has escaped X inactivation (Figure S11).
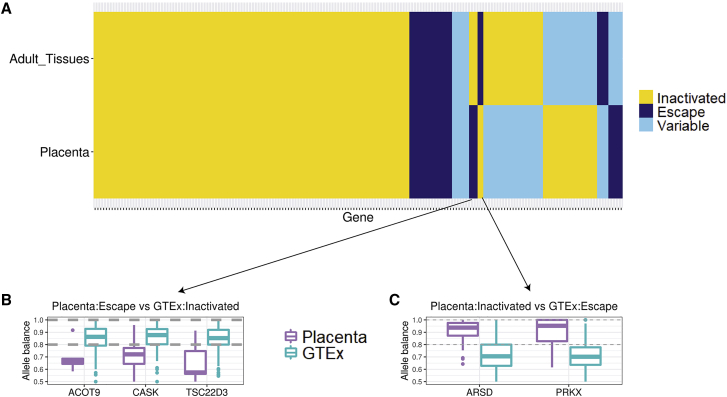
We found similarities and differences in gene-specific escape and inactivation in the human placenta when compared with the adult tissues. We used 525 skewed adult tissue samples in the GTEx dataset (out of 4,958 total samples) and 52 skewed samples in the placenta dataset (out of 58 total samples) to categorize XCI status (see material and methods). A gene is considered for this analysis if there are at least five informative samples (i.e., an informative sample is defined as having at least one heterozygous and expressed variant for that gene) in both the placenta dataset and the GTEx dataset. We found 186 such genes. Out of these 186 genes, 132 genes (71%) show the same pattern of escape or inactivation between the adult tissues and the placenta dataset (Figure 4A). Specifically, 111 genes are inactivated, 15 genes escape, and 6 genes show variable escape between the adult tissues and the placenta. We also observed differences between the adult tissues and the placenta. We found three genes that escape in the placenta but are inactivated in adult tissues: ACOT9, CASK, and TSC22D3 (Figure 4B). We found two genes that are inactivated in the placenta but escape in adult tissues: ARSD and PRKX (Figure 4C). We examined the mapping quality and total RNA read counts of the variants in these genes to confirm that these observations are not artifacts (Figure S12). These results suggest that, while the majority of genes on the X chromosome show consistent XCI between the placenta and adult tissues, a subset exhibits opposite patterns that could be attributed to the unique nature of the placenta tissue, which is an embryonic tissue.
Figure 4.
Gene-specific escape comparison between the placenta and adult tissues
(A) A heatmap showing genes that are inactivated in both the placenta and adult GTEx tissues (yellow), genes that escape X chromosome inactivation in both the placenta and adult GTEx tissues (dark blue), and genes that exhibit variable escape across individuals in both the placenta and adult GTEx tissues (light blue).
(B and C) Boxplots showing median allele balance across all samples in the placenta (purple) and adult tissues in GTEx (blue) for the three genes that show evidence for escaping XCI in the placenta but being inactivated in the adult GTEx tissues (B) and for the two genes that show evidence for being inactivated in the placenta but escaping in the adult GTEx tissues (C). Each point of the box plot is the median allele balance computed across variants for that gene (see material and methods).
Finally, for a subset of our placenta samples, we also had access to maternal exome samples and used these to assess whether the silenced sample was maternal or paternal in origin and observed that most inactivated X chromosomes were paternal in origin (Table S7). In 18/30 placentas, we also sequenced the whole exome of the decidua, allowing us to determine whether the maternal or paternal X chromosome is inactivated (see material and methods). In 8/18 placenta samples where both extraction sites show the same inactivated X chromosome, it is the paternal X that is silenced in both (Table S7). In 3/18 placenta samples where one extraction site shows skewed X inactivation while the other extraction site shows both X chromosomes being expressed, in two of these cases, the paternal X is silenced (Table S7). In 7/18 placenta samples the two extraction sites necessarily show the opposite X chromosome being activated, either maternal or paternal (Table S7).
Discussion
We observed that XCI in the human placenta is organized into large patches of maternal or paternal X expression. We utilized whole-exome and whole-transcriptome sequence data to analyze allele-specific expression across female placentas and found evidence that the human placenta exhibits large patches of maternal or paternal X chromosome expression. While patchy XCI in the human placenta has been observed in humans previously, these prior studies rely on a few SNPs and a few genes in a limited number of samples.24,45 For example, Looijenga et al.45 examined two samples from each of the nine female (46, XX) full-term placenta samples, using methylation in a single gene, androgen receptor, as a readout, they found that three placentas exhibited predominantly maternal X inactivation, one exhibited paternal X inactivation, and the remaining five exhibited both maternal and paternal alleles. In a different study, with at least five informative SNPs used to infer X inactivation status in a single site collected from 22 placentas, the authors similarly observe some placentas have maternal X inactivation, some have paternal X inactivation, and others show biallelic expression across the X-linked SNPs.24 From this, the authors proposed that the human placenta has large patches of maternal or paternal X inactivation. These studies typically relied on a single gene, as in Looijenga et al.,45 or when these studies did not include multiple samplings from the same placenta, the number of samples is small (i.e., three) as in Moreira de Mello et al.24 With improved sampling (i.e., sampling two independent locations from each placenta, from opposing quadrants) and genome-wide sequencing, we confirm that patterns of XCI are predominantly patchy in the human placenta.
We found distinct patterns of XCI between the placenta and adult tissues in humans: while most placenta samples (∼90%) exhibit skewed expression, only about 11% of samples from human adult tissues exhibit skewed expression. This is consistent with previous methylation-based analyses that observed skewed inactivation across different developmental layers of the placenta,25 suggesting that each villus tree is clonally derived from a few precursor cells and that there is little cell migration across placental regions. This is in contrast to adult tissues, where we observe very little skewing, suggesting more cell migration during development, though both observations are contingent on the size of the tissue sample analyzed in bulk, where samples with more cells are more likely to capture both X chromosomes being expressed. Per our reading of the sample collection in GTEx (see material and methods), the bulk sample sizes were approximately the same size at collection (aiming for 10 mm × 10 mm × 4 mm) compared with the placenta samples collected here (10–20 mm cubed); however, it is not clear what size of this sample was sequenced by GTEx, compared with what was sequenced here (2 mm cubed was sequenced per placenta sample). It is possible that a larger bulk sample was sequenced by the GTEx consortium, resulting in the signal for biallelic expression across most adult tissues.
Although we found high concordance in genes that are inactivated and genes that escape between the placenta samples and adult tissues (Figure 4), we found a subset of genes with unique XCI status in the placenta. Specifically, we found five genes that show patterns that are unique to the placenta: ACOT9, CASK, and TSC22D3 are inactivated in adult tissues but escape XCI in the placenta (Figure 4B). ACOT9 has been associated with syndromic X-linked intellectual disability Turner Type,46 but the function of ACOT9 in the placenta is unclear. CASK is critical in brain development post-natally,47 as well as prenatal brain development in both males and females, but with varying phenotypes,48 potentially due to its escape from inactivation (biallelic expression) in females compared with males. TSC22D3 is an immunity-related gene. Syrett al.49 observed that TSC22D3 is overexpressed in females compared with males in T cells of patients with systemic lupus erythematosus. In contrast to the previous three genes, ARSD and PRKX, both genes in the pseudoautosomal region, escape XCI in adult tissues but are inactivated consistently across regions in the placenta (Figure 4C). Larson et al.50 observed that ARSD is inactivated in at least one ovarian tumor sample, highlighting a potential parallel between the placenta and tumor development.50 PRKX has been previously reported to escape in adult fibroblasts and lymphoblasts.51 Curiously, PRKX is also involved in fetal kidney development,52 and so perhaps its silencing is important for dosage during development, but further studies are needed to determine whether there are sex differences in PRKX expression in the placenta. We hypothesize that these five genes could be further studied for a potential role in pregnancy and pregnancy complications.
We additionally show that sex differences in gene expression are not sufficient on their own to determine whether a gene escapes inactivation or not (Figure S11). This highlights the importance of understanding both inactivation status and expression differences between the sexes. Gong et al.22 used female-biased gene expression as a proxy for identifying potential genes that escape XCI in the placenta. Twenty-eight of forty-seven potential escape genes identified by Gong et al.22 were reported previously as being inactivated or unknown. Of these, 3/28 genes exhibit variable escape in our placenta data (MBTPS2, SMS, and PIN4) and only 1/28 genes exhibit evidence for escaping (CXorf36). This means the other 24/28 show female-biased expression but no evidence of escape in the placenta. Thus, it will be critical for studies interested in studying XCI to assess allele-specific expression explicitly.
There has also been interest in whether the maternal or paternal X chromosome is preferentially inactivated. In mice, the paternal X is preferentially inactivated in extraembryonic tissues,53 potentially due to unique methylation of XIST in sperm compared with eggs resulting.54 Further, the paternal X is preferentially inactivated in rat yolk sac.7 However, as we show here, approximately half of our samples show patchiness of XCI, with both parents’ X chromosomes being expressed in one quadrant or the other. This is consistent with observed randomness of X inactivation in the placenta in horses as well.12 That said, for the subset of samples where only one parent’s X chromosome is expressed in both quadrants, we curiously observe that, overwhelmingly, it is the paternal X chromosome that is inactivated.
Future work should also focus on confirming whether certain genes that show a variable pattern of inactivation across the two quadrants studied here are indeed variable using spatial transcriptomic or single-cell analyses. Single-cell data for the placenta have been generated (e.g., Pique-Regi et al.,55 Vento-Tormo et al.,56 and Ashary et al.57) that could potentially be used for future analyses of this inactivation heterogeneity. Unfortunately, the samples used in this study were collected in a manner inconsistent with using them for single-cell isolation.
In conclusion, we confirmed that XCI in the human placenta is skewed but patchy and conducted allele-specific expression analyses in adult tissues to show that, in contrast to the placenta, most adult samples show mosaic inactivation in bulk samples. Using the subset of adult tissues that do show skewed inactivation, we identified a subset of genes showing patterns of XCI that are unique to the placenta that should be further investigated for their roles in pregnancy and placentation. We further report that sex differences in expression are not sufficient to predict X inactivation and that, where there is bias (only one parent’s X chromosome being inactivated across the two samples), this tends to be the parental X chromosome.
Acknowledgments
We acknowledge Research Computing at Arizona State University for providing high-performance computing and storage resources that have contributed to the research results reported within this paper (http://www.researchcomputing.asu.edu). We would also like to thank Heini Natri and Wilson lab members for helpful feedback on the manuscript. This work was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health grant R35GM124827 to M.A.W. This research was supported by The Yale University Reproductive Sciences Data and Specimen Biorepository, HIC#1309012696, a component of the Department of Obstetrics, Gynecology & Reproductive Sciences, Yale School of Medicine, New Haven, CT. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under award number F31HD101252 to K.C.O. K.C.O. was additionally supported by ARCS Spetzler Scholar.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100121.
Supplemental information
Data and code availability
The datasets generated during this study are available via controlled access: phs002240.v1.p1 NIGMS Sex Differences Placentas. The code generated during this study is available at https://github.com/SexChrLab/Placenta_XCI.
References
- 1.Natri H., Garcia A.R., Buetow K.H., Trumble B.C., Wilson M.A. The pregnancy pickle: evolved immune compensation due to pregnancy underlies sex differences in human diseases. Trends Genet. 2019;35:478–488. doi: 10.1016/j.tig.2019.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilson Sayres M.A., Makova K.D. Gene survival and death on the human Y chromosome. Mol. Biol. Evol. 2013;30:781–787. doi: 10.1093/molbev/mss267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takagi N., Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 5.Huynh K.D., Lee J.T. Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr. Opin. Cell Biol. 2001;13:690–697. doi: 10.1016/s0955-0674(00)00272-6. [DOI] [PubMed] [Google Scholar]
- 6.Huynh K.D., Lee J.T. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat. Rev. Genet. 2005;6:410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- 7.Wake N., Takagi N., Sasaki M. Non-random inactivation of X chromosome in the rat yolk sac. Nature. 1976;262:580–581. doi: 10.1038/262580a0. [DOI] [PubMed] [Google Scholar]
- 8.Xue F., Tian X.C., Du F., Kubota C., Taneja M., Dinnyes A., Dai Y., Levine H., Pereira L.V., Yang X. Aberrant patterns of X chromosome inactivation in bovine clones. Nat. Genet. 2002;31:216–220. doi: 10.1038/ng900. [DOI] [PubMed] [Google Scholar]
- 9.Richardson B.J., Czuppon A.B., Sharman G.B. Inheritance of glucose-6-phosphate dehydrogenase variation in kangaroos. Nat. New Biol. 1971;230:154–155. doi: 10.1038/newbio230154a0. [DOI] [PubMed] [Google Scholar]
- 10.Cooper D.W., Johnston P.G., Vandeberg J.L., Robinson E.S. X-chromosome inactivation in marsupials. Aust. J. Zool. 1989;37:411–417. doi: 10.1071/zo9890411. [DOI] [Google Scholar]
- 11.Al Nadaf S., Waters P.D., Koina E., Deakin J.E., Jordan K.S., Graves J.A. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11:R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Miller D.C., Clark A.G., Antczak D.F. Random X inactivation in the mule and horse placenta. Genome Res. 2012;22:1855–1863. doi: 10.1101/gr.138487.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrel L., Brown C.J. When the Lyon(ized chromosome) roars: ongoing expression from an inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20160355. doi: 10.1098/rstb.2016.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 15.Cotton A.M., Price E.M., Jones M.J., Balaton B.P., Kobor M.S., Brown C.J. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 2015;24:1528–1539. doi: 10.1093/hmg/ddu564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berletch J.B., Ma W., Yang F., Shendure J., Noble W.S., Disteche C.M., Deng X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tukiainen T., Villani A.-C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James J.L., Carter A.M., Chamley L.W. Human placentation from nidation to 5 weeks of gestation. Part I: what do we know about formative placental development following implantation? Placenta. 2012;33:327–334. doi: 10.1016/j.placenta.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 19.Turco M.Y., Moffett A. Development of the human placenta. Development. 2019;146:dev163428. doi: 10.1242/dev.163428. [DOI] [PubMed] [Google Scholar]
- 20.Gude N.M., Roberts C.T., Kalionis B., King R.G. Growth and function of the normal human placenta. Thromb. Res. 2004;114:397–407. doi: 10.1016/j.thromres.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 21.Rathbun K.M., Hildebrand J.P. StatPearls. StatPearls Publishing; 2019. Placenta abnormalities. [Google Scholar]
- 22.Gong S., Sovio U., Aye I.L., Gaccioli F., Dopierala J., Johnson M.D., Wood A.M., Cook E., Jenkins B.J., Koulman A., et al. Placental polyamine metabolism differs by fetal sex, fetal growth restriction, and preeclampsia. JCI Insight. 2018;3 doi: 10.1172/jci.insight.120723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui Y., Chen Q., Sun X. Association of skewed X chromosome inactivation and idiopathic recurrent spontaneous abortion: a systematic review and meta-analysis. Reprod. Biomed. Online. 2015;31:140–148. doi: 10.1016/j.rbmo.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Moreira de Mello J.C., Araújo É.S. S.d., Stabellini R., Fraga A.M., Souza J.E. S.d., Sumita D.R., Camargo A.A., Pereira L.V. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peñaherrera M.S., Jiang R., Avila L., Yuen R.K.C., Brown C.J., Robinson W.P. Patterns of placental development evaluated by X chromosome inactivation profiling provide a basis to evaluate the origin of epigenetic variation. Hum. Reprod. 2012;27:1745–1753. doi: 10.1093/humrep/des072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slavney A., Arbiza L., Clark A.G., Keinan A. Strong constraint on human genes escaping X-inactivation is modulated by their expression level and breadth in both sexes. Mol. Biol. Evol. 2016;33:384–393. doi: 10.1093/molbev/msv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konwar C., Del Gobbo G., Yuan V., Robinson W.P. Considerations when processing and interpreting genomics data of the placenta. Placenta. 2019;84:57–62. doi: 10.1016/j.placenta.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available online. [Google Scholar]
- 30.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushnell B. Lawrence Berkeley National Lab. (LBNL); 2014. BBMap: A Fast, Accurate, Splice-Aware Aligner. [Google Scholar]
- 32.Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. Arxiv. 2013 doi: 10.48550/arXiv.1303.3997. [DOI] [Google Scholar]
- 33.Webster T.H., Couse M., Grande B.M., Karlins E., Phung T.N., Richmond P.A., Whitford W., Wilson M.A. Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. GigaScience. 2019;8 doi: 10.1093/gigascience/giz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picard Tools - By Broad Institute. http://broadinstitute.github.io/picard/
- 36.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., Weir B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olney K.C., Brotman S.M., Valverde-Vesling V., Andrews J., Wilson M.A. Aligning RNA-Seq reads to a sex chromosome complement informed reference genome increases ability to detect sex differences in gene expression. Biol. Sex Differ. 2020;11:42. doi: 10.1186/s13293-020-00312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim D., Langmead B., Salzberg S. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barnett D.W., Garrison E.K., Quinlan A.R., Strömberg M.P., Marth G.T. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castel S.E., Levy-Moonshine A., Mohammadi P., Banks E., Lappalainen T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 2015;16:195. doi: 10.1186/s13059-015-0762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Human Genome Overview - Genome Reference Consortium.
- 45.Looijenga L.H., Gillis A.J., Verkerk A.J., van Putten W.L., Oosterhuis J.W. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am. J. Hum. Genet. 1999;64:1445–1452. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016;54:1–30. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava S., McMillan R., Willis J., Clark H., Chavan V., Liang C., Zhang H., Hulver M., Mukherjee K. X-linked intellectual disability gene CASK regulates postnatal brain growth in a non-cell autonomous manner. Acta Neuropathol. Commun. 2016;4:30. doi: 10.1186/s40478-016-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moog U., Bierhals T., Brand K., Bautsch J., Biskup S., Brune T., Denecke J., de Die-Smulders C.E., Evers C., Hempel M., et al. Phenotypic and molecular insights into CASK-related disorders in males. Orphanet J. Rare Dis. 2015;10:44. doi: 10.1186/s13023-015-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Syrett C.M., Paneru B., Sandoval-Heglund D., Wang J., Banerjee S., Sindhava V., Behrens E.M., Atchison M., Anguera M.C. Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. JCI Insight. 2019;4 doi: 10.1172/jci.insight.126751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larson N.B., Fogarty Z.C., Larson M.C., Kalli K.R., Lawrenson K., Gayther S., Fridley B.L., Goode E.L., Winham S.J. An integrative approach to assess X-chromosome inactivation using allele-specific expression with applications to epithelial ovarian cancer. Genet. Epidemiol. 2017;41:898–914. doi: 10.1002/gepi.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wainer Katsir K., Linial M. Human genes escaping X-inactivation revealed by single cell expression data. BMC Genom. 2019;20:201. doi: 10.1186/s12864-019-5507-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li X., Li H.-P., Amsler K., Hyink D., Wilson P.D., Burrow C.R. PRKX, a phylogenetically and functionally distinct cAMP-dependent protein kinase, activates renal epithelial cell migration and morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9260–9265. doi: 10.1073/pnas.132051799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harper M.I., Fosten M., Monk M. Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J. Embryol. Exp. Morphol. 1982;67:127–135. doi: 10.1242/dev.67.1.127. [DOI] [PubMed] [Google Scholar]
- 54.Zuccotti M., Monk M. Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nat. Genet. 1995;9:316–320. doi: 10.1038/ng0395-316. [DOI] [PubMed] [Google Scholar]
- 55.Pique-Regi R., Romero R., Tarca A.L., Sendler E.D., Xu Y., Garcia-Flores V., Leng Y., Luca F., Hassan S.S., Gomez-Lopez N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife. 2019;8 doi: 10.7554/elife.52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A., et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashary N., Bhide A., Chakraborty P., Colaco S., Mishra A., Chhabria K., Jolly M.K., Modi D. Single-cell RNA-seq identifies cell subsets in human placenta that highly expresses factors driving pathogenesis of SARS-CoV-2. Front. Cell Dev. Biol. 2020;8:783. doi: 10.3389/fcell.2020.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available via controlled access: phs002240.v1.p1 NIGMS Sex Differences Placentas. The code generated during this study is available at https://github.com/SexChrLab/Placenta_XCI.