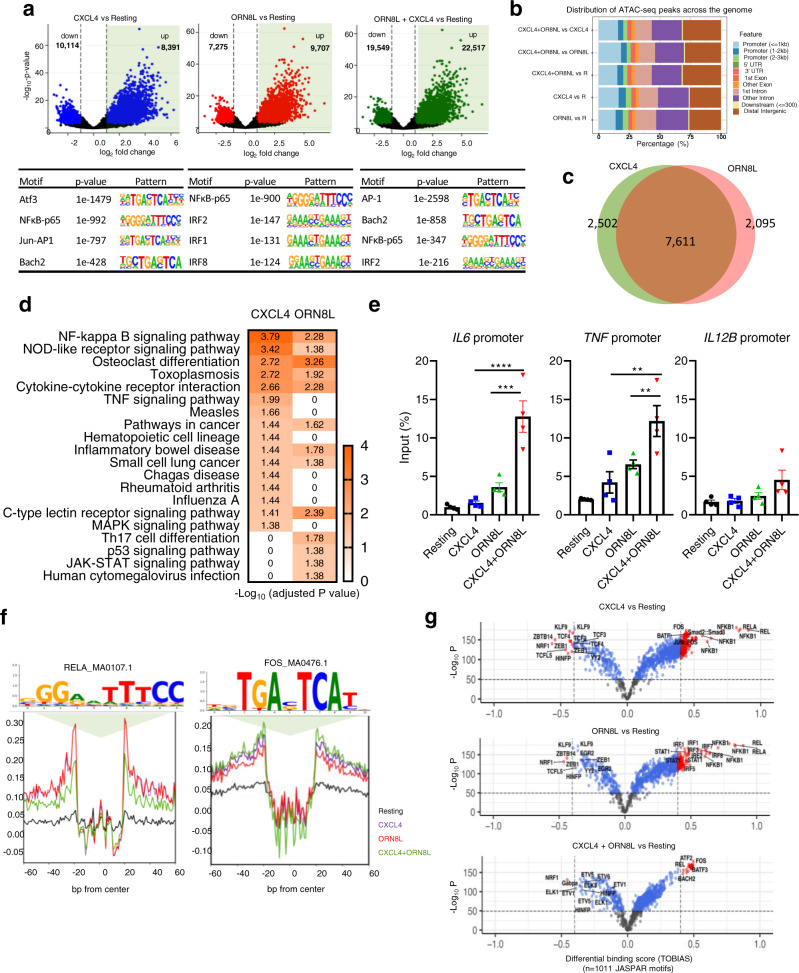
Fig. 4. ATACseq analysis of induction of open chromatin regions (OCRs) by CXCL4 and TLR8.
a Volcano plots (upper panels) showing differentially induced (right) or suppressed (left) peaks after individual or combined treatment with CXCL4 and ORN8L compared to untreated resting (R) monocytes. DESeq2 Wald test statistic was used to rank the peaks wherein only peaks with Benjamini–Hochberg corrected false discovery rate (FDR) values <0.05 and rank scores above or below 2.5 were considered significant and were chosen for downstream analysis. Lower panels show motif enrichment by HOMER known motif analysis of open chromatin regions. b Chart representing the relative distribution of ATACseq peak coordinates across the genome. c Venn diagram showing overlapping and distinct ATACseq peaks. d KEGG pathway enrichment analysis of genes associated with OCRs. Color scale represents adjusted p-value (<0.05), fold change ≥3. e FAIRE assays of the IL6, TNF, and IL12B promoter regions 6 h after CXCL4 and/or TLR8 stimulation. n = 3 independent experiments. f Visualization of RELA_MA0107.1 and FOS_MA0476.1 motif footprints by TOBIAS BINDetect algorithm. g Volcano plot depiction of differential binding analysis of n = 1011 JASPAR motifs by TOBIAS using BINDetect algorithm. The x-axis represents differential binding score and y-axis −log10 p-value. Motifs with significant binding are highlighted in blue and significant motifs with highest differential binding scores in red. ATACseq analysis (a–d, f, g) is based on 3 independent experiments. Data are depicted as mean ± SEM (e). ****p ≤ 0.0001; ***p ≤ 0.001; **p ≤ 0.01 by one-way ANOVA (e). Source data are provided as a Source data file.