Fig. 2. High-resolution cryo-EM structure of Ad5-3M virus, possessing compound mutations in hexon HVR1 and HVR7 loops and penton base RGD loop.
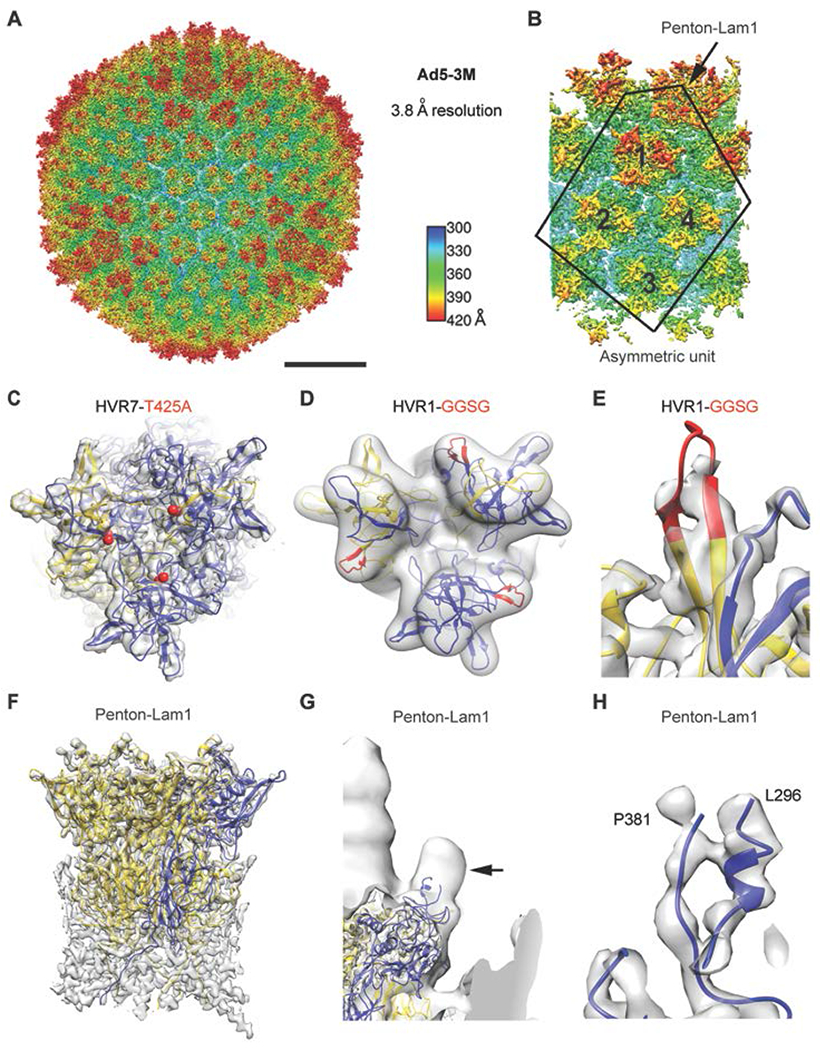
(A) Full Ad5-3M capsid structure at 3.8 Å resolution displayed with radial coloring. Scale bar, 250 Å. (B) Enlarged view of the icosahedral asymmetric unit (outlined), which includes one subunit of penton base with a laminin-α1 derived integrin interacting motif and 4 hexon trimers with mutations in HVR1 and HVR7 loops. (C) Top view of one Ad5-3M hexon trimer with the sites of the Thr-425-Ala (T425A) HVR7 loop point mutation depicted with red spheres. One hexon subunit is in gold, two hexon subunits are in blue, and the cryo-EM density is in transparent gray. (D) Ad5-3M hexon density filtered to 5-Å resolution superimposed with hexon coordinates including the modeled HVR1 loop containing the Gly-Gly-Ser-Gly (GGSG) sequence (red). (E) Enlarged view of Ad5-3M HVR1 loop model shown with unfiltered cryo-EM density. (F) Side view of the Ad5-3M penton base pentamer (one subunit in blue, four subunits in gold). (G) Ad5-3M penton base density filtered to 15-Å resolution reveals a protrusion at the base of integrin interacting loop (arrow). Density for the fiber, which protrudes from the center of the penton base, is observed at the left edge of the panel. (H) Enlarged view of ordered residues at either end of the Ad5-3M penton base integrin interacting loop shown with unfiltered cryo-EM density.
