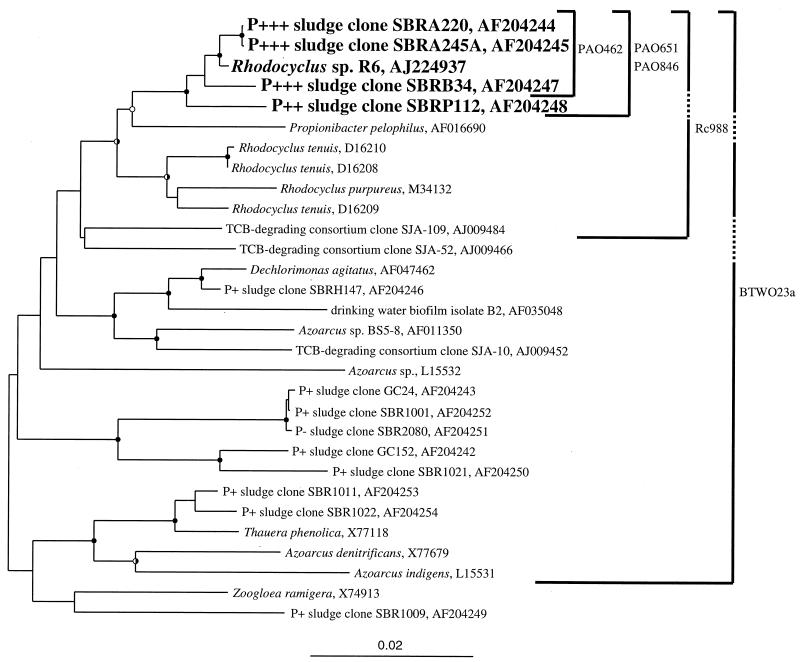
FIG. 3.
Phylogenetic tree of near-complete 16S rDNA sequences obtained from sludges A, B, P, SBRH, SBR1, SBR2, and GC (Gold Coast, Queensland, Australia) determined in this study and sequences from publicly accessible databases. Rubrivivax gelatinosus (D16213) was used as the outgroup in analyses but is not shown in the tree. Evolutionary distance and parsimonious analyses were carried out in PAUP∗ employing 2,000 bootstrap resamplings. Closed circles at nodes indicate >75% bootstrap support from both analyses; open circles indicate 50 to 75% from both analyses; half-shaded circles are for analyses where one algorithm gave >75% bootstrap support and the other gave 50 to 75%. The coding indicates that the clone came from a hyper-P-removing sludge (ca. 15% P in the sludge) (P+++), a good P-removing sludge (P++), a fair P-removing sludge (P+), or a non-P-removing sludge (P−). The specificity of the published β-2 Proteobacteria probe (BTWO23a) and those of probes designed in this work (PAO probes and Rc988) are shown by solid lines. Dashed lines indicate that the probe does not have 100% identity with the indicated sequence. For example, Rc988 has one mismatch (at position 1009) with the SBRP112 sequence. In addition to specifically targeting the sequences indicated in the tree, probe BTWO23a also targets (with no mismatches) members of the β-3 and γ Proteobacteria, Iodobacter, Chromobacterium, Chromatium spp., and a green nonsulfur division clone, OPB9. The scale indicates 0.02 nucleotide change per nucleotide position. TCB, trichlorobenzene.