Abstract
Background
Immune-stimulatory agents, like agonists of the innate immune receptor RIG-I, are currently tested in clinical trials as an intratumoral treatment option for patients with unresectable melanoma, aiming to enhance anti-tumor T cell responses. Switching of melanoma toward a dedifferentiated cell state has recently been linked to T cell and therapy resistance. It remains to be determined whether RIG-I agonists affect melanoma differentiation, potentially leading to T cell resistance.
Methods
Patient metastases-derived melanoma cell lines were treated with the synthetic RIG-I agonist 3pRNA, and effects on tumor cell survival, phenotype and differentiation were determined. Transcriptomic data sets from cell lines and metastases were analyzed for associations between RIG-I (DDX58) and melanoma differentiation markers and used to define signaling pathways involved in RIG-I-driven dedifferentiation. The impact of 3pRNA-induced melanoma dedifferentiation on CD8 T cell activation was studied in autologous tumor T cell models.
Results
RIG-I activation by 3pRNA induced apoptosis in a subpopulation of melanoma cells, while the majority of tumor cells switched into a non-proliferative cell state. Those persisters displayed a dedifferentiated cell phenotype, marked by downregulation of the melanocytic lineage transcription factor MITF and its target genes, including melanoma differentiation antigens (MDA). Transition into the MITFlow/MDAlow cell state was JAK-dependent, with some cells acquiring nerve growth factor receptor expression. MITFlow/MDAlow persisters switched back to the proliferative differentiated cell state when RIG-I signaling declined. Consistent with our in vitro findings, an association between melanoma dedifferentiation and high RIG-I (DDX58) levels was detected in transcriptomic data from patient metastases. Notably, despite their dedifferentiated cell phenotype, 3pRNA-induced MITFlow/MDAlow persisters were still efficiently targeted by autologous CD8 tumor-infiltrating T lymphocytes (TILs).
Conclusions
Our results demonstrate that RIG-I signaling in melanoma cells drives a transient phenotypic switch toward a non-proliferative dedifferentiated persister cell state. Despite their dedifferentiation, those persisters are highly immunogenic and sensitive toward autologous TILs, challenging the concept of melanoma dedifferentiation as a general indicator of T cell resistance. In sum, our findings support the application of RIG-I agonists as a therapeutic tool for the generation of long-term clinical benefit in non-resectable melanoma.
Keywords: melanoma; tumor escape; immunotherapy; lymphocytes, tumor-infiltrating
WHAT IS ALREADY KNOWN ON THIS TOPIC
Human melanoma is highly plastic and can adapt to environmental stresses by transition into dedifferentiated cell states, which have repeatedly been associated with T cell resistance.
Synthetic agonists of the innate pattern recognition receptor RIG-I are administered intratumorally to improve anti-tumor T cell responses, although knowledge about their impact on the differentiation state of melanoma cells is lacking.
WHAT THIS STUDY ADDS
Here, we demonstrate that in response to RIG-I activation subpopulations of melanoma cells switch into a non-proliferative dedifferentiated persister cell state, from which they escape when RIG-I signaling declines.
Despite their JAK-STAT-dependent dedifferentiation, those persisters can be efficiently targeted by autologous tumor-infiltrating lymphocytes.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE AND/OR POLICY
RIG-I-induced melanoma dedifferentiation is not a barrier to effective anti-tumor T cell responses, supporting the application of RIG-I agonists in tumor therapy and suggesting that melanoma dedifferentiation is not a general but rather context-dependent indicator of T cell resistance.
Background
Immune checkpoint blockade (ICB) has revolutionized the treatment of advanced melanoma, but clinical responses are still limited to a subgroup of patients. Cytotoxic CD8 T cells are central anti-tumor effectors in ICB. Accordingly, ICB resistance has been associated with metastases lacking CD8 T cell infiltrates and expressing low levels of HLA class I (HLA-I) molecules presenting antigens to CD8 T cells.1–3 To overcome resistance and improve clinical responses, innate pattern recognition receptors (PRRs) attracted attention as targets for combination ICB therapies.
Intratumoral application of PRR agonists can induce tumor cell death and orchestrate local innate and adaptive anti-tumor immune responses, potentially translating into systemic immunity.4–6 This has also been demonstrated for mimetics of viral double-stranded RNA (dsRNA) binding to the ubiquitous cytosolic immune receptor retinoic acid-inducible gene I (RIG-I).7 8 Activation of RIG-I by its synthetic agonist 5′-triphosphate dsRNA (3pRNA) triggers the production of type I interferons (IFN-I) and T cell-attracting chemokines in melanoma cells.2 8 Moreover, PRR signaling enhances melanoma cell immunogenicity by upregulation of the HLA-I antigen processing and presentation machinery (HLA-I APM) and has the capacity to overcome tumor cell-intrinsic resistance to T cells.2 9 There is accumulating evidence also from preclinical studies that PRR signaling synergizes with ICB in boosting the anti-tumor activity of CD8 T cells in melanoma.2 10–12 These findings and the clinical observation that patients with RIG-I (DDX58)-high melanomas show improved survival2 11 provided a strong rational for clinical trials combining intratumoral injection of synthetic RIG-I ligands with anti-PD-1/-L1 antibodies.
During therapy, tumor cells are exposed to various environmental pressures that melanoma cells can adapt to and escape from via phenotype switching.13 14 For instance, under prolonged exposure to proinflammatory cytokines or inhibitors of oncogenic signaling pathways (MAPK inhibitors, MAPKi), differentiated melanoma cells can stably switch into diverse dedifferentiated cell states.15–17 Common to these cell states is the downregulation of the melanocytic lineage Microphthalmia-associated transcription factor (MITF) and its target gene network.15–17 Of note, certain dedifferentiated cell states are characterized by the upregulation of specific phenotypic markers, such as receptor tyrosine kinase AXL or nerve growth factor receptor (NGFR/CD271),14 18–21 the latter recently described as a regulator of melanoma phenotype switching.18
Melanoma dedifferentiation is of highest clinical relevance, as it has repeatedly been observed in tumor biopsies derived from patients resistant to targeted inhibitor therapy or immunotherapy.3 22–24 Therapy resistance of dedifferentiated melanomas has been attributed, at least partly, to the lower immunogenicity of dedifferentiated cells, lacking expression of melanoma differentiation antigens (MDAs), such as Melan-A and tyrosinase.15–17 22
Our previous studies demonstrated that short-term RIG-I activation in melanoma cells (up to 24 hours) immediately enhances their sensitivity toward autologous CD8 T cells.2 However, tumor-T cell contacts might be delayed, in particular, when RIG-I agonists are injected into immune-cold lesions. Here, we demonstrate that under prolonged RIG-I signaling, melanoma cells transiently switch into a dedifferentiated but highly immunogenic persister cell state, still sensitive to autologous CD8 T cells. Our findings suggest that despite induction of melanoma dedifferentiation, intratumoral injection of RIG-I agonists could be a valuable strategy for improvement of melanoma therapy.
Materials and methods
Patient samples
Tumor tissues were collected after written informed consent. Studies on human material were approved by the institutional review board. Melanoma cell lines (Ma-Mel-47, Ma-Mel-61a, Ma-Mel-63a, Ma-Mel-86c, UKE-Mel-164a) were established from metastatic lesions, as previously described.16 25 26 Cells were confirmed to be mycoplasma-free in monthly intervals and authenticated by genetic profiling on genomic DNA at the Institute for Forensic Medicine (University Hospital Essen) using the AmpFLSTR-Profiler Plus kit (Applied Biosystems). Melanoma cell lines were cultured in RPMI1640 medium supplemented with 10% (v/v) fetal calf serum (FCS) and penicillin/streptomycin except for UKE-Mel-164a, maintained in Dulbecco's Modified Eagle Medium (DMEM) medium supplemented with 10% FCS, penicillin/streptomycin, 1% non-essential amino acids and 1% sodium pyruvate. All cells were grown at 5% CO2, 37°C.
Transfection of RIG-I agonists
The here used 3pRNA and non-stimulatory control (Ctrl) RNA were described previously.2 High molecular weight poly(I:C) was purchased from Invivogen. Tumor cells were transfected with 100–200 ng/mL 3pRNA, Ctrl RNA or 50–100 ng/mL poly(I:C) using Lipofectamine 2000 (Invitrogen). All transfections were performed according to the manufacturer’s instructions. Serum addition (Ma-Mel cells) or medium exchange (UKE-Mel-164a) was carried out 6 hours post-transfection. Cells were incubated at 5% CO2, 37°C, and subjected to further analyses at the indicated time points.
Transfection efficiency was estimated using a fluorescence-conjugated microRNA mimic (Dharmacon), resembling 3pRNA in structure (double-stranded) and size. Cells were transfected with 100–200 ng/mL microRNA according to the 3pRNA transfection protocol. After 24 hours, cells were harvested, washed at least three times, and subjected to fluorescence measurement applying the Gallios flow cytometer (Beckman Coulter). Data analysis was carried out using Kaluza software (Beckman Coulter).
Inhibitor and cytokine treatment
Inhibitor treatment
Cells were seeded and transfected with 3pRNA or Ctrl RNA as described above. Six hours post-3pRNA-transfection, cells were treated with 0.5 or 2 µM JAK1/2 inhibitor (JAKi, Ruxolitinib; Selleckchem) for 3 days.
Cytokine treatment
Cells were seeded and 24 hours later exposed to interferon-γ (IFN-γ) (500 IU/mL; Imukin, Boehringer Ingelheim) or tumor necrosis factor-α (TNF-α) (100 ng/mL; Boehringer Ingelheim) for the indicated time period. Control cells were left untreated.
Annexin V–propidium iodide staining
On day 3 post-transfection, cells were harvested and washed with phosphate-buffered saline (PBS). For staining, pellets were resuspended in binding buffer (10 mM HEPES, pH 7.4; 140 mM NaCl; 2.5 mM CaCl2) containing Annexin V-APC (BD Pharmingen) and propidium iodide (PI; BD Pharmingen) and incubated at room temperature in the absence of light for 15 min. Afterward, PBS was added, and cells were immediately measured. Measurement and data processing were performed using the Gallios flow cytometer and Kaluza software, respectively (Beckman Coulter).
Western blot
Tumor cell lysates were obtained using 1x Cell Lysis Buffer (Cell Signaling). About 15 µg protein/sample was separated on 8%–12% sodium dodecyl sulfate (SDS)-polyacrylamide gels, electroblotted onto nitrocellulose membranes, and probed with the following primary antibodies at 4°C overnight: anti-human rabbit antibodies anti-RIG-I (clone D14G6), anti-STAT1 (clone D1K9Y), anti-phospho (Tyr701) STAT1 (clone 58D6), anti-IRF1 (clone D5E4, XP), anti-CDK2 (clone 78B2), anti-p21 Waf1/Cip1 (clone 12D1), anti-p27 Kip (clone D69C12, XP), anti-AXL (clone C89E7), anti-c-jun (clone 60A8), anti-phosphoSer73 c-jun (clone D47G9), anti-FRA1 (clone D80B4) and anti-GAPDH (clone 14C10) were purchased from Cell Signaling. Anti-human mouse antibodies used: anti-tyrosinase (clone T311, Santa Cruz), anti-MITF (clone C5, Sigma-Aldrich), anti-MART1/Melan-A (clone M2-7C10, Zytomed). After washing, membranes were incubated with the appropriate secondary antibody conjugated with horseradish peroxidase. Antibody binding was visualized using enhanced chemiluminescence system (Thermo Scientific). In all experiments, GAPDH served as a loading control.
Quantitative real-time PCR (qPCR)
Total mRNA isolation, reverse transcription and TaqMan-based real-time PCR were performed as previously described.2 26 The amount of specific mRNA was normalized to endogenous GAPDH (Ma-Mel-47) or RPLP0 (Ma-Mel-61a) mRNA levels. Due to the instability of different house keeping genes in 3pRNA-transfected Ma-Mel-61a cells, specific mRNA levels of 3pRNA-treated Ma-Mel-61a cells were normalized to RPLP0 mRNA levels of Ctrl RNA-transfected cells. Fold change in relative mRNA expression was calculated by normalizing the expression of treated cells to the corresponding controls.
Microscopy
Cell morphology and density were documented by light microscopy (Olympus CKX41) using the Olympus CellSens Standard software.
Real-time proliferation assay (xCELLigence)
Background was measured with 50 µL medium per well in an E-Plate 16 (Roche). Then 3×103 or 5×103 cells/well were seeded in 150 µL additional volume. Cell attachment was monitored with RTCA SP (Roche) instrument and the RTCA Software V.1.2 (Roche) every 10 min. After 16–20 hours, melanoma cells were transfected with 100–200 ng/mL 3pRNA or Ctrl RNA and incubated for 10–15 days at 5% CO2, 37°C. As additional controls, cells were treated with Lipofectamine 2000 only or left untransfected. Six hours post-transfection, 10% FCS was added to Ma-Mel-86c and Ma-Mel-47 cells, the medium was exchanged entirely for UKE-Mel-164a cells. All experiments were carried out as triplicates. Changes in the electric impedance were given as a dimensionless cell index value derived from relative impedance changes corresponding to cellular coverage of the electrode sensors. The cell index was first normalized to the baseline impedance value of the background and then to the time point of transfection.
Flow cytometry
Melanoma cells were harvested at the indicated time points after transfection and once washed with PBS, followed by staining with live/dead stain kit (Invitrogen) and incubation at room temperature in the absence of light for 30 min. After washing, cells were split up for intracellular and surface staining. For surface staining, cells were incubated with the following antibodies: anti-AXL-SB436 (clone DS7HAXL, Invitrogen), anti-HLA-ABC-APC (clone W6/32, Invitrogen), anti-CD271(NGFR)-PE (clone ME20.4, BioLegend) and fixed with 4% paraformaldehyde. For intracellular staining, cells were fixed and permeabilized using Fixation/Permeabilization Kit (eBioscience) and stained with the following antibodies: anti-Melan-A-FITC (clone A103, Santa Cruz) and anti-CD271(NGFR)-APC/Fire750 (clone ME20.4, BioLegend). Fixed samples were measured using the Gallios flow cytometer (Beckman Coulter). Autofluorescence was determined by unstained samples, and Kaluza software (Beckman Coulter) was used for data analysis. Fold change in relative mean fluorescence intensity (MFI) was calculated by normalizing the MFI of treated cells to the corresponding controls.
Cell sorting
Ma-Mel-47 were transfected with 3pRNA or Ctrl RNA. On day 3 post-transfection, cells were stained with anti-CD271(NGFR)-APC/Fire750 (clone ME20.4, BioLegend). Stained 3pRNA-transfected cells were separated into NGFRhigh and NGFRlow subpopulations applying the BD FACSAria III cell sorter. Aliquots of sorted cells were analyzed for their differentiation status by intracellular Melan-A staining. The remaining cells were cultured until regrowth, followed by analyses of intracellular Melan-A and NGFR expression levels via flow cytometry, as described above.
CFSE proliferation assay
Ma-Mel-61a cells were transfected with 3pRNA or Ctrl RNA. On day 3 post-transfection, cells were stained with 10 µM CellTrace carboxyfluorescein diacetate succinimidyl ester (CFSE) Cell Proliferation Kit (Invitrogen) according to the manufacturer’s instructions. CFSE-labled cells were harvested and co-stained for intracellular NGFR at the indicated time points followed by flow cytometry analysis.
Expansion of autologous CD8 T cells
Tumor-reactive bulk CD8 T cells from peripheral blood of patient Ma-Mel-61 and CD8 TILs from patient Ma-Mel-86 were expanded following previously described protocols.16 26 Expansion of tumor-reactive tumor-infiltrating lymphocytes (TILs) from patient UKE-Mel-164 was carried out as follows: Single-cell suspension from fresh tumor tissue was obtained using GentleMACS dissociation kit (Miltenyi Biotec). CD8 T cells were isolated using anti-CD8 MicroBeads (Miltenyi Biotec). Isolated T cells (1×106) were co-cultured in 24-well culture plates with irradiated autologous melanoma cells (1×105) in AIM-V medium (Gibco/BRL) supplemented with 10% (v/v) human serum at 5% CO2, 37°C. On day 3, recombinant human IL-2 (250 IU/mL) was added. In weekly intervals, 106 T cells were restimulated with 105 irradiated melanoma cells.
T cell activation assay
Intracellular cytokine staining was performed to determine the activation of CD8 T cells by 3pRNA-transfected and poly(I:C)-transfected autologous melanoma cells. The T cell-stimulatory capacity of 3pRNA, poly(I:C) or Ctrl RNA-treated melanoma cells was determined 24 hours and/or 3 days after transfection. For long-term 3pRNA or Ctrl RNA treatment, melanoma cells were transfected twice (days 0 and 6) and subjected to T cells 24 hours later (day 7). Briefly, T cells were stimulated with indicated autologous melanoma cells (1:1 ratio) for 4 hours in the presence of Brefeldin A (10 µg/mL; Sigma-Aldrich) at 5% CO2, 37°C. Afterward, cells were fixed and permeabilized using Fixation/Permeabilization Kit (eBioscience) and stained with an antibody cocktail containing anti-CD3-BV421 (clone UCHTI, BioLegend); anti-CD8-APC/Cy7 (clone SK1, BioLegend), anti-IFN-γ-PE (clone B27, BioLegend) and anti-TNF-α-PE/Cy7 (clone Mab11, BioLegend). For cell analysis and data processing, the Gallios flow cytometer and Kaluza software were used, respectively (Beckman Coulter).
Public RNA-seq data analyses
RNA sequencing (RNA-seq) data from human melanoma cell lines (Tsoi data set27) and patient samples (Gide cohort,28 Liu cohort29) were analyzed by dividing the cohorts into equally sized groups of samples with low and high expression of RIG-I (DDX58). Groups were compared for expression of MLANA, MITF, DCT, TYR, and CD8A. For the Gide cohort, gene expression was quantified from fastq files in transcripts per million mapped reads (TPM) per gene using kallisto.30 The annotation of transcripts to genes was used as provided by kallisto in the Homo_sapiens.GRCh38.96.gtf from the isoform estimates. The data on gene expression from the other cohorts were used as published. Immune infiltrates were quantified applying the TIMER webtool31 using the TPM expression values of the Gide and Liu cohorts with the SKCM cancer type selected.32
Enrichment analysis
For the enrichment analysis RIG-I (DDX58), co-regulated genes were determined using Spearman correlation in RNA-seq data of the Tsoi melanoma cell line cohort.27 The conservative correction for the number of tests yielded Bonferroni-corrected q-values, and FDR-corrected q-values were computed by applying p.adjust() in R using the FDR method. Genes were then sorted by significance. The top 50 correlated genes were queried in enrichment analysis using the Enrichr webtool33 at https://maayanlab.cloud/Enrichr/. The resulting enrichments were considered statistically significant if p values were less than 0.05.
Statistics
Experimental data from three independent experiments are depicted as means plus SE of the mean (±SEM). For comparison of experimental groups, the two-tailed paired t-test was applied using GraphPad Prism V.8 software (GraphPad). For group comparisons of RNA-seq data, the two-sided Wilcoxon rank test was applied. Experimental groups were considered to be significantly different at levels of p value less than 0.05.
Results
3pRNA-treated melanoma cells persist in a transient non-proliferative cell state
To study the effects of RIG-I signaling on melanoma cell states, we transfected various tumor cell lines, established from patient metastases, with 3pRNA or Ctrl RNA. Tumor cells were treated once, according to clinical trial protocols applying PRR agonists intratumorally in 3-week intervals (NCT03739138, NCT02828098). Intact RIG-I signaling was confirmed by elevated RIG-I protein expression in 3pRNA-treated melanoma cells (figure 1A) and cytotoxic effects of the highly efficient transfection procedure were excluded (online supplemental figure S1A–D).
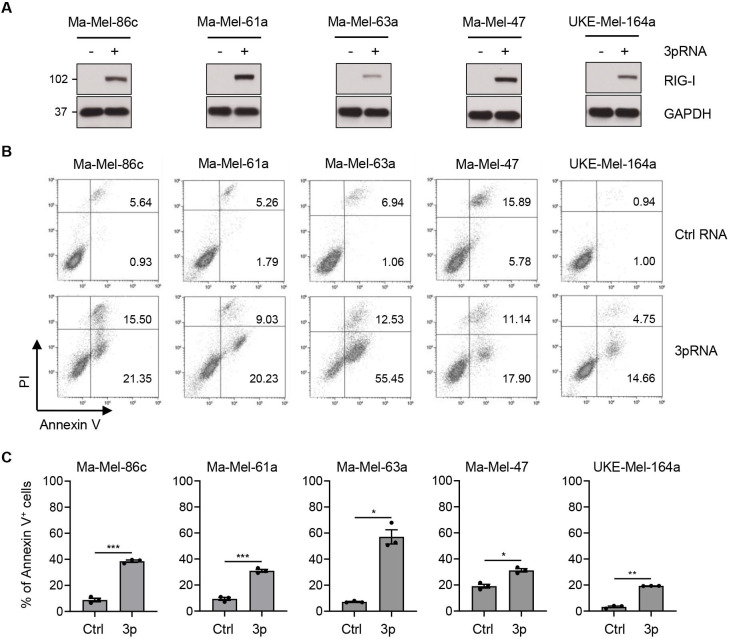
Figure 1.
Enhanced melanoma cell death in response to 3pRNA treatment. (A–C) Melanoma cells were transfected with 3pRNA (+) or Ctrl RNA (-) and subjected to further analyses on day 3 post-transfection. (A) RIG-I expression analyzed by western blot. GAPDH, loading control. Representative data from three independent experiments. (B, C) Cells stained for Annexin V/PI and analyzed by flow cytometry. (B) Representative dot plots, (C) percentage of Annexin V-positive (Annexin V+) cells shown as mean±SEM from three independent experiments. Significantly different experimental groups: *p<0.05, **p<0.01, ***p<0.005 by two-tailed paired t-test. Ctrl, control; PI, propidium iodide; RIG-I, retinoic acid-inducible gene I.
jitc-2021-003863supp001.pdf (5.1MB, pdf)
Activation of RIG-I in tumor cells has been demonstrated to trigger apoptosis.34–36 As shown in figure 1B by Annexin V-PI staining, we detected significantly enhanced cell death in all 3pRNA-treated melanoma cell lines (figure 1B, C). But, except for cell line Ma-Mel-63a, the majority of transfected cells (60%–80%) remained viable (figure 1C).
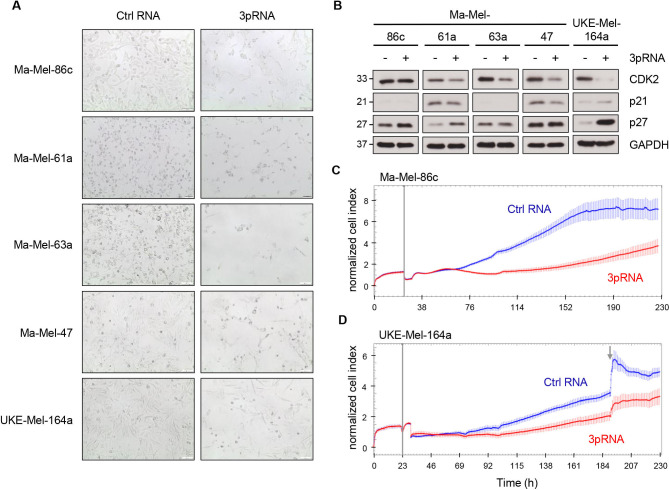
Analyzing the cells that persisted upon RIG-I activation, we recognized a more spindle-like mesenchymal cell shape of 3pRNA-treated melanoma cells compared with control cells (figure 2A). Moreover, persisters switched into a non-proliferative cell state, indicated by downregulation of the cell cycle regulator CDK2 in four out of five 3pRNA-treated cell lines and upregulation of the CDK inhibitor p27 in Ma-Mel-61a, Ma-Mel-86c, and UKE-Mel-164a cells (figure 2B). We confirmed the strong inhibitory effect of RIG-I activation on Ma-Mel-86c and UKE-Mel-164a cells in real-time proliferation assays applying an impedance-based technology (figure 2C, D; online supplemental figure S2A, B). Strikingly, on day 4 to day 5 after 3pRNA transfection, melanoma cells resumed proliferation. Overall, these data demonstrated that prolonged RIG-I signaling in melanoma cells induced a transient non-proliferative cell state.
Figure 2.
Induction of a transient non-proliferative melanoma cell state upon RIG-I activation. (A, B) Melanoma cells were transfected with 3pRNA (+) or Ctrl RNA (-) and subjected to further analyses on day 3 post-transfection. (A) Microscopic analyses of cell morphology and density (×10 magnification). Representative images from three independent experiments. (B) Expression of cell cycle regulators analyzed by western blot. GAPDH, loading control. Representative data from three independent experiments. (C, D) Real-time proliferation of cell lines Ma-Mel-86c (C) and UKE-Mel-164a (D) after 3pRNA (red) or Ctrl RNA (blue) transfection. Vertical gray lines indicate time point of transfection; gray arrow shows medium exchange. Representative data from three independent experiments. Ctrl, control; RIG-I, retinoic acid-inducible gene I.
Persisting melanoma cells acquire a dedifferentiated phenotype
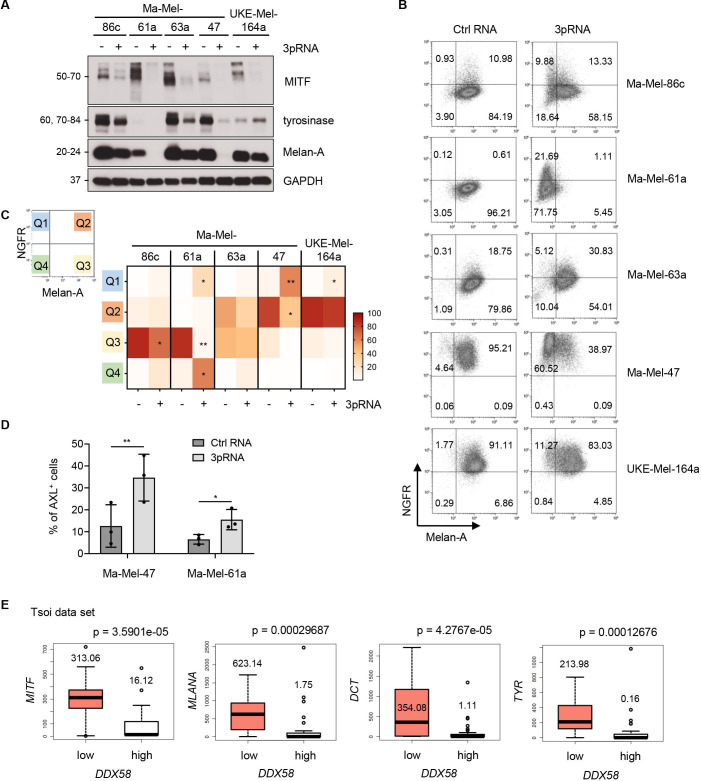
The lineage transcription factor MITF functions as a driver of melanoma cell proliferation.37 As shown by western blot in figure 3A, 3pRNA-induced non-proliferative persisters strongly downregulated the expression of MITF and its targets Melan-A and tyrosinase. Some cell lines (Ma-Mel-61a, Ma-Mel-47) appeared to even switch off differentiation marker expression. Considering that loss in melanoma differentiation can be accompanied by an upregulation of dedifferentiation markers, such as AXL or NGFR,14 21 38 we examined the expression of Melan-A, AXL, and NGFR in 3pRNA-treated and Ctrl RNA-treated cells by flow cytometry. Consistent with the western blot analyses, a decrease in Melan-A was measured also by flow cytometry in all cell lines tested, with Ma-Mel-47 lacking protein expression in the majority of cells and Ma-Mel-61a displaying a nearly complete Melan-A loss. Among the 3pRNA-induced Melan-A-negative cells, we detected subpopulations of cells upregulating NGFR or AXL. An increase in the NGFRhigh/Melan-Aneg fraction (Q1) was observed in all investigated melanoma cell lines (figure 3B, C), while an enrichment of AXL-positive cells was detected only in two cases (figure 3D; online supplemental figure S3A, B). Thus, prolonged RIG-I signaling induced a non-proliferative dedifferentiated melanoma cell state.
Figure 3.
Dedifferentiation of 3pRNA-induced persister cells. (A–D) Melanoma cells were transfected with 3pRNA (+) or Ctrl RNA (-) and subjected to further analyses on day 3 post-transfection. (A) Expression of differentiation markers analyzed by western blot. GAPDH, loading control. Representative data of three independent experiments. (B, C) Single-cell phenotype profiling by flow cytometry. (B) Representative dot plots of NGFR and Melan-A expression. Numbers indicate the percentage of specific cell populations. Quadrants defined based on autofluorescence of unstained cells. (C) Heatmap of the quadrant‘s median of three independent experiments. (D) Surface expression of AXL measured by flow cytometry. Percentage of AXL-positive (AXL+) cells shown as mean±SEM of three independent experiments. Significantly different experimental groups: *p<0.05, **p<0.01 by two-tailed paired t-test. (E) Analysis of Tsoi transcriptomic data set,27 comparing gene expression in patient-derived melanoma cell lines grouped by RIG-Ilow (DDX58low) and RIG-Ihigh (DDX58high) (n=27 vs n=27; threshold: 7 TPM). P values calculated by two-sided Wilcoxon rank tests. Ctrl, control; NGFR, nerve growth factor receptor; RIG-I, retinoic acid-inducible gene I; TPM, transcripts per million mapped reads.
To verify our finding that melanoma differentiation and RIG-I levels are inversely correlated, we analyzed public transcriptomic data from 53 patient-derived melanoma cell lines (Tsoi data set).27 Melanoma cell lines were divided into RIG-I (DDX58) low and high expression groups and compared for levels of MITF and its targets MLANA, DCT, and TYR. As shown in figure 3E, expression of each marker was significantly decreased in the RIG-Ihigh (DDX58high) compared with RIG-Ilow (DDX58low) group, in line with our findings that elevated RIG-I protein expression (figure 1A) in melanoma cells is associated with dedifferentiation.
RIG-I-induced melanoma dedifferentiation involves JAK-STAT signaling
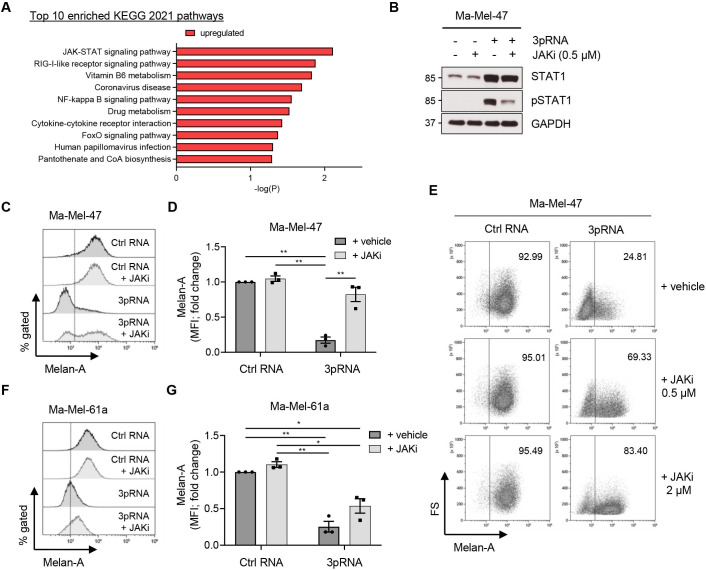
To decipher the mechanisms underlying 3pRNA-induced dedifferentiation, we performed pathway analysis on the Tsoi transcriptome data set27 and found genes involved in IFN and JAK/STAT signaling strongly enriched in RIG-Ihigh (DDX58high) melanoma cells (figure 4A; online supplemental table S1A, B and figure S4). Considering also our previous observations of IFN-I expression by 3pRNA-treated tumor cells,2 we postulated the involvement of JAK-STAT signaling in melanoma dedifferentiation. To analyze this, we selected melanoma models Ma-Mel-47 and Ma-Mel-61a, showing strongest RIG-I-induced dedifferentiation in terms of Melan-A downregulation (figure 3A–C), and treated those cells with 3pRNA in the presence or absence of the JAK inhibitor (JAKi) Ruxolitinib. As shown for Ma-Mel-47 in figure 4B, JAK-STAT signaling, triggered in response to RIG-I activation, was strongly decreased in the presence of Ruxolitinib. The inhibitor also blocked Melan-A downregulation by 3pRNA in a concentration-dependent manner (figure 4C–E). Similar effects of JAKi were observed for 3pRNA-treated Ma-Mel-61a cells (figure 4F, G), indicating an involvement of JAK-STAT signaling in RIG-I-induced melanoma dedifferentiation.
Figure 4.
JAK-dependent dedifferentiation of 3pRNA-induced persisters. (A) KEGG database pathways enriched for genes co-regulated with RIG-I (DDX58) mRNA in the Tsoi transcriptomic data set.27 (B) Ma-Mel-47 cells were transfected with 3pRNA or Ctrl RNA, incubated with or without 0.5 µM JAKi (Ruxolitinib) for 24 hours. JAK-STAT signaling analyzed by western blot. GAPDH, loading control. Representative data of two independent experiments. (C, D) Ma-Mel-47 cells were transfected with 3pRNA or Ctrl RNA, incubated with or without 0.5 µM JAKi (Ruxolitinib), and subjected to further analyses on day 3 post-transfection. Intracellular Melan-A expression determined by flow cytometry. (C) Representative histograms with dotted line representing unstained control and (D) fold change of MFI given as mean±SEM of three independent experiments. (E) Melan-A expression in Ma-Mel-47 after 3pRNA transfection and incubation with different JAKi concentrations for 3 days; representative dot plots of two or three independent experiments. (F, G) Ma-Mel-61a cells were transfected with 3pRNA or Ctrl RNA, incubated with or without 0.5 µM JAKi (Ruxolitinib) for 3 days, and subjected to further analyses. Intracellular Melan-A expression determined by flow cytometry. (F) Representative histograms with dotted line representing unstained control and (G) fold change of MFI given as mean±SEM of three independent experiments. Significantly different experimental groups: **p<0.01 by two-tailed paired t-test. Ctrl, control; MFI, mean fluorescence intensity.
jitc-2021-003863supp002.xlsx (265.3KB, xlsx)
In addition to 3pRNA, JAK-STAT pathway activation and tumor cell dedifferentiation were induced also by transfection of poly(I:C), demonstrating that distinct synthetic RIG-I agonists trigger the same molecular and phenotypic alterations in melanoma cells (online supplemental figure S5A–D).
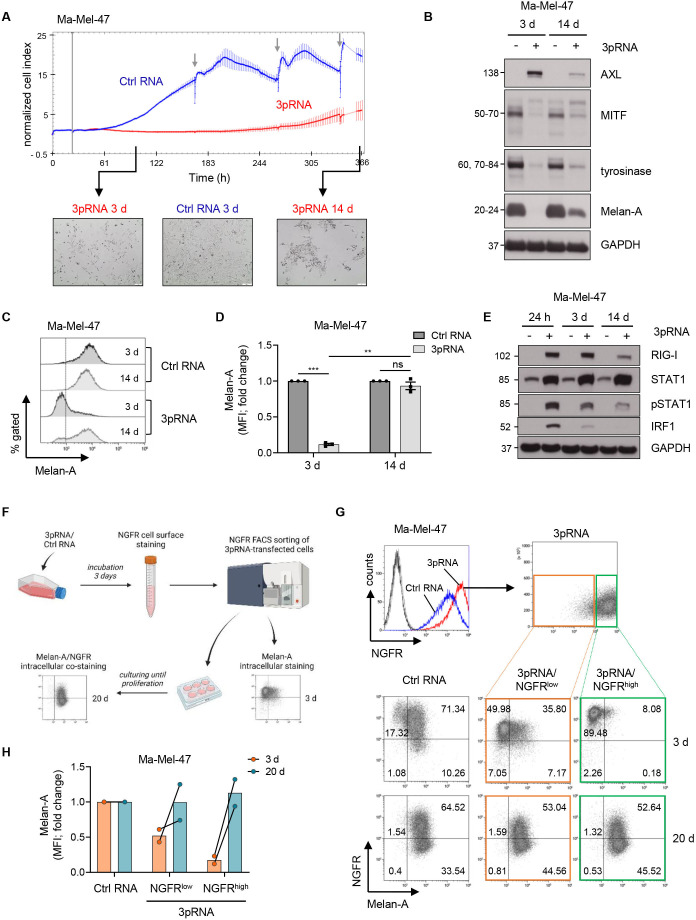
Decline in RIG-I signaling reverses melanoma phenotype switching
To address the transient nature of dedifferentiation, we harvested persisting Ma-Mel-47 cells on day 3 and day 14 post-3pRNA-transfection. Time points were selected based on real-time cell viability measurement (figure 5A), with day 3 corresponding to the non-proliferative cell state and day 14 to the regrowth phase. As shown in figure 5B, on day 3 of RIG-I activation, the majority of non-proliferative Ma-Mel-47 persisters lost Melan-A expression. In contrast, when cells regained their proliferative capacity (day 14), the Melan-A-positive phenotype dominated (figure 5B–D). Consistent with a back-switch toward a differentiated phenotype, proliferative Ma-Mel-47 persisters showed a decrease in NGFR and AXL expression (figure 5B; online supplemental figure S6E) and downregulation in JAK-STAT signaling (figure 5E). Similar time-dependent changes in the expression of differentiation and dedifferentiation markers were observed for 3pRNA-treated Ma-Mel-61a cells (online supplemental figure S6A–E).
Figure 5.
Transient melanoma cell dedifferentiation upon RIG-I activation. (A–E) Ma-Mel-47 cells were transfected once with 3pRNA or Ctrl RNA and analyzed on day 3 (3 d) and 14 (14 d) post-transfection. (A, top) Real-time proliferation of Ma-Mel-47 after 3pRNA (red) or Ctrl RNA (blue) transfection. Vertical gray line indicates time point of transfection; gray arrows show medium exchange. Representative data of three independent experiments. (A, bottom) Morphology and cell density monitored microscopically (×10 magnification). Representative images of three independent experiments. (B) Differentiation status determined by the expression of indicated proteins. GAPDH, loading control. Representative data of three independent experiments. (C, D) Melan-A expression determined by flow cytometry. (C) Representative histograms with dotted line representing unstained control and (D) fold change of MFI given as mean±SEM of three independent experiments. (E) RIG-I expression and JAK-STAT pathway activation analyzed by western blot. GAPDH, loading control. Representative data of three independent experiments. (F–H) 3pRNA-transfected Ma-Mel-47 were sorted for NGFR expression on day 3 post-transfection and cultured until reproliferation (20 d). (F) Scheme of workflow. Illustration created with BioRender.com. (G, top) NGFR surface expression before cell sorting on day 3 post-transfection. (G, bottom) Melan-A and NGFR coexpression in melanoma cells analyzed by flow cytometry directly after cell sorting (3 d) and at the time point of reproliferation on day 20 post-transfection (20 d). Representative dot plots with percentage of cells from two independent experiments. (H) Quantification of Melan-A expression given as fold change of MFI from two independent experiments. Linked points indicate results from the same experiment. Significantly different experimental groups: **p<0.01; ***p<0.005 by two-tailed paired t-test. Ctrl, control; MFI, mean fluorescence intensity; MITF, Microphthalmia-associated transcription factor; NGFR, nerve growth factor receptor; RIG-I, retinoic acid-inducible gene I.
To stress the reversibility of RIG-I-driven phenotype switching and demonstrate that even highly dedifferentiated persisters can switch back to a differentiated proliferative cell state, we sorted 3pRNA-induced NGFRhigh Ma-Mel-47 cells from NGFRlow cells (day 3 post-transfection) (figure 5F, G). Of the sorted NGFRhigh cells, more than 91% showed a dedifferentiated Melan-A-negative phenotype compared with 57% of the NGFRlow population (figure 5G, H). Both the NGFRlow and NGFRhigh population resumed proliferation and switched back toward a differentiated phenotype similar to control cells (day 20 post-transfection) (figure 5G, H).
Notably, transition back to proliferation was observed also for 3pRNA-treated Ma-Mel-61a persisters, which consisted of up to 93% dedifferentiated cells on day 3 post-transfection (figure 3B; online supplemental figure S6E). To exclude that proliferative cells were exclusively derived from the small subpopulation of Ma-Mel-61a cells that preserved their differentiated phenotype under 3pRNA treatment (figure 3B; online supplemental figure S6E), we monitored the reproliferation of CFSE-labeled persister in a short-term setting. Bulk adherent persister cells were stained with CFSE on day 3 post-3pRNA-transfection and fluorescence intensity was measured on day 3, day 6 and day 8. Flow cytometry analysis revealed a time-dependent gradual loss of CFSE signal intensity in total cells, associated with a reexpression of Melan-A (online supplemental figure S7), clearly demonstrating that even highly dedifferentiated persisters switched back toward a differentiated cell state and resumed proliferation.
Taken together, our findings suggest that RIG-I signaling in melanoma cells induces a transient non-proliferative dedifferentiated cell state via JAK-STAT pathway activation that tumor cells escape from when RIG-I signaling declines.
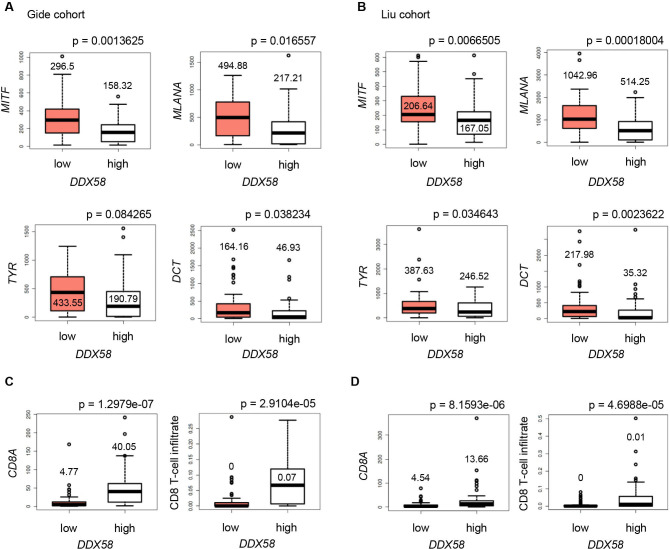
Dedifferentiated RIG-Ihigh tumors show enhanced T cell infiltration
To address the clinical relevance of our findings, we studied RIG-I (DDX58) and differentiation marker expression in public transcriptomic data sets from melanoma tissues. Analysis of two independent patient cohorts revealed a significant downregulation of differentiation genes (MITF, MLANA, TYR, DCT) in RIG-Ihigh (DDX58high) compared with RIG-Ilow (DDX58low) tumors (figure 6A, B). Moreover, RIG-Ihigh (DDX58high) melanomas showed elevated CD8A expression (figure 6C, D). Applying the TIMER webtool, we estimated the CD8 T cell infiltrates based on the transcriptomic data and found them increased in RIG-Ihigh (DDX58high) lesions (figure 6C, D), consistent with the elevated CD8A expression. In summary, the tissue expression data strengthened our in vitro findings, and the enhanced infiltration of RIG-Ihigh (DDX58high) melanomas by CD8 T cells led us to ask for the reactivity of CD8 T cells toward 3pRNA-induced dedifferentiated persister cells.
Figure 6.
RIG-I (DDX58) expression in melanoma biopsies correlates with dedifferentiation and T cell infiltration. Expression of melanoma differentiation markers MITF, MLANA, TYR, and DCT (A, B), and T cell marker CD8A (C, D, left) in RIG-Ilow (DDX58low) and RIG-Ihigh (DDX58high) expression groups from published RNA-seq data sets. (C, D, right) Calculated CD8 T cell infiltrates in RIG-Ilow (DDX58low) and RIG-Ihigh (DDX58high) expression groups. Data sets of the Gide cohort28 (A, C, n=45 vs n=46; threshold: 11.1 TPM) and Liu cohort29 (B, D; n=60 vs n=61; threshold: 7.7 TPM). P values from two-sided Wilcoxon rank tests. MITF, Microphthalmia-associated transcription factor; RIG-I, retinoic acid-inducible gene I; TPM, transcripts per million mapped reads.
CD8 T cells efficiently respond toward RIG-I-induced dedifferentiated persisters
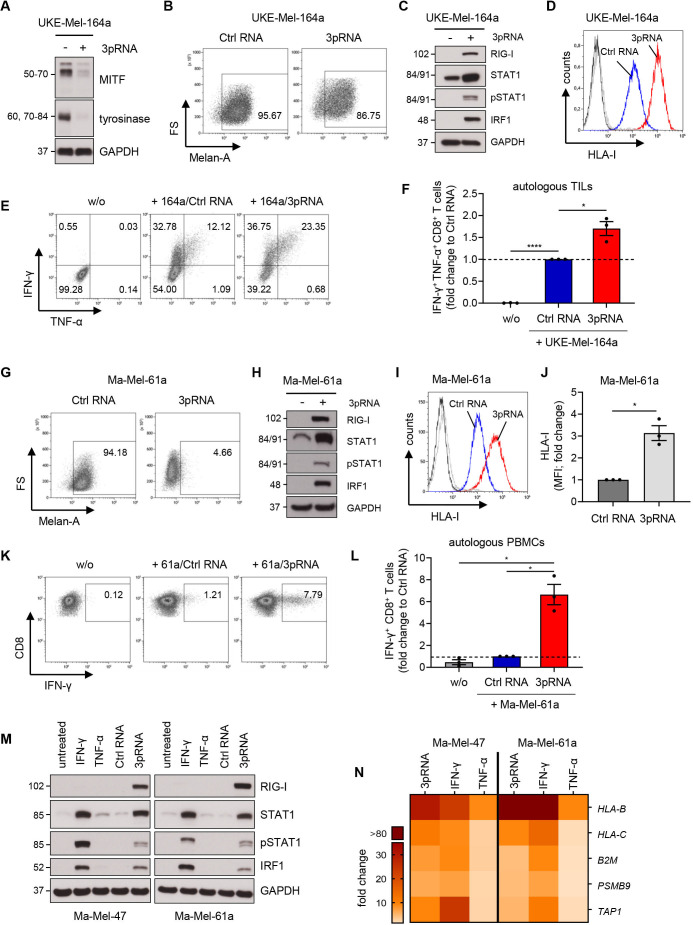
Recently, we demonstrated impaired recognition of MAPKi-induced dedifferentiated melanoma cells by CD8 T cells.16 17 To study T cell responses toward 3pRNA-induced persisters, we took advantage of various autologous CD8 T cell-melanoma cell models.
In patient model UKE-Mel-164, we analyzed the recognition of 3pRNA-induced persisters by autologous TILs. To enhance RIG-I signaling and melanoma dedifferentiation, tumor cells were transfected twice with 3pRNA. Compared with single transfection (figure 3A), we detected a stronger downregulation of the differentiation antigen tyrosinase (figure 7A), but still, the majority of melanoma cells retained Melan-A expression (figure 7B). RIG-I signaling also triggered JAK-STAT pathway activation (figure 7C) and upregulation of HLA-I surface molecules (figure 7D). Strikingly, the amount of TILs producing IFN-γ and/or TNF-α was strongly increased in the presence of 3pRNA-induced persisters compared with Ctrl RNA-treated tumor cells (figure 7E, F).
Figure 7.
Strong CD8 T cell reactivity toward 3pRNA-induced dedifferentiated persisters. (A, B and D, F) UKE-Mel-164a cells were transfected twice with 3pRNA (+) or Ctrl RNA (-) and subjected to further analyses on day 7 post-transfection. (A) Differentiation status of UKE-Mel-164a determined by the expression of indicated proteins. GAPDH, loading control. Representative data from three independent experiments. (B) Melan-A expression determined by flow cytometry. Representative histogram from three independent experiments. (C) RIG-I expression and JAK-STAT pathway activation analyzed 24 hours post-transfection by western blot. GAPDH, loading control. Representative data from three independent experiments. (D) HLA-I cell surface expression measured by flow cytometry. Representative histogram from three independent experiments. (E, F) Activation of autologous TILs by UKE-Mel-164a cells transfected with 3pRNA or Ctrl RNA analyzed by intracellular cytokine staining in flow cytometry. (E) Representative dot plot and (F) quantification of IFN-γ+ TNF-α+ CD8+ TILs. Fold change given as mean±SEM from three independent experiments. (G–L) Ma-Mel-61a melanoma cells were transfected with 3pRNA (+) or Ctrl RNA (-) and subjected to further analyses on day 1 (H) or day 3 (G, I–L) post-transfection. (G) Melan-A expression analyzed by flow cytometry. Representative data from three independent experiments. (H) RIG-I expression and JAK-STAT pathway activation analyzed on day 1 post-transfection by western blot. GAPDH, loading control. Representative data from three independent experiments. (I, J) HLA-I cell surface expression measured by flow cytometry. (I) Representative histogram and (J) fold change of MFI given as mean±SEM from three independent experiments. (K, L) Activation of autologous T cells by Ma-Mel-61a cells transfected with 3pRNA or Ctrl RNA analyzed by intracellular IFN-γ staining in flow cytometry. (K) Representative dot plot and (L) quantification of IFN-γ+ CD8+ T cells. Fold change given as mean±SEM from two independent experiments. (M) RIG-I and JAK/STAT pathway activation analyzed 24 hours post-transfection with 3pRNA or Ctrl RNA or post-treatment with IFN-γ or TNF-α by western blot. GAPDH, loading control. Representative data from two independent experiments. (N) Relative mRNA expression of genes involved in antigen processing and presentation in Ma-Mel-47 and Ma-Mel-61a cells on day 3 post-3pRNA transfection or post-cytokine treatment. Fold change given as mean from two independent experiments. Significantly different experimental groups: *p<0.05; ****p<0.001 by two-tailed paired t-test. Ctrl, control; IFN, interferon; MITF, Microphthalmia-associated transcription factor; PBMC, peripheral blood mononuclear cell; RIG-I, retinoic acid-inducible gene I; TIL, tumor-infiltrating T lymphocyte; TNF, tumor necrosis factor, w/o: without target cells.
With Ma-Mel-86c, we studied a second melanoma model, showing stronger dedifferentiation in response to RIG-I activation than UKE-Mel-164a (figure 3A; online supplemental figure S8A, B). We confirmed JAK-STAT pathway activation and increased HLA-I expression in response to RIG-I signaling (online supplemental figure S8C–E). 3pRNA treatment of Ma-Mel-86c cells triggered enhanced activation of autologous TILs at early time points (24 hours) (online supplemental figure S8F, H), in line with our previous study.2 In contrast, persister cells showed T cell-stimulatory capacity similar to control cells (online supplemental figure S8G, H).
So far, the functional data suggested that T cell responses toward dedifferentiated persisters were not impaired but even enhanced. However, persisters from both patient models still contained subpopulations of partially differentiated cells, potentially contributing to T cell activation. To overcome this limitation, we finally incorporated the melanoma model Ma-Mel-61 in our study. Tumor cells from this patient showed a complete loss of MITF, tyrosinase, and Melan-A protein expression on day 3 post-3pRNA-transfection (figures 3A and 7G), concurrent to JAK-STAT pathway activation and HLA-I upregulation (figure 7H–J). Despite total dedifferentiation, 3pRNA-induced persisters strongly activated autologous CD8 T cells in contrast to Ctrl RNA-treated melanoma cells (figure 7K, L). Thus, persisting Ma-Mel-61a cells showed enhanced immunogenicity and triggered T cell activation by antigens other than the lost differentiation antigens. Similar to 3pRNA-induced persisters, also poly(I:C)-transfected Ma-Mel-61a cells displayed improved T cell-stimulatory capacity (online supplemental figure S9A–C).
3pRNA-induced persisters upregulate the HLA-I antigen presenting machinery similar to IFNγ-treated cells
Our previous study demonstrated increased expression of the HLA-I APM by short-term 3pRNA-treated melanoma cells,2 suggesting that HLA-I APM upregulation could contribute also to the enhanced T cell-stimulatory capacity of 3pRNA-induced persisters. To estimate the level of HLA-I APM upregulation, we compared the expression of specific APM components in 3pRNA-induced persisters to IFN-γ- and TNF-α-treated melanoma cells. IFN-γ is known as potent HLA-I APM inducer and both IFN-γ and TNF-α have been described to trigger melanoma dedifferentiation.15 22 39 In fact, each agent (3pRNA, IFN-γ, TNF-α) downregulated MITF and its target genes in our melanoma models (online supplemental figure S10A, B). But, only 3pRNA and IFN-γ potently triggered JAK/STAT pathway activation linked to the induction of HLA-I APM genes, including HLA-B, HLA-C, B2M and PSMB9 involved in antigen peptide presentation and processing, respectively (figure 7M, N). In line with this observation, HLA-I upregulation was strongest on 3pRNA-induced dedifferentiated NGFRhigh persisters (online supplemental figure S11A–C), indicating their potent antigen-presenting capacity.
Overall, our data from the different melanoma models clearly demonstrate that, despite inducing dedifferentiation, RIG-I activation in melanoma cells preserves or even enhances the immunogenicity of persisting melanoma cells, enabling efficient anti-tumor T cell responses.
Discussion
The capacity of RIG-I agonists to enhance innate and adaptive anti-tumor immune responses has been demonstrated in various preclinical studies,2 11 40 41 leading to their implementation in clinical trial protocols. RIG-I ligands are applied intratumorally in a carrier-complexed format, thereby acting on stromal and tumor cells. Previous studies demonstrated that uptake of 3pRNA in tumor cells can induce mitochondrial apoptosis in vitro and in vivo.34–36 Accordingly, in this study, we observed an increase in cell death in 3pRNA-treated patient-derived melanoma cell lines. However, the majority of melanoma cells remained viable under 3pRNA treatment and persisted in a non-proliferative state. RIG-I-induced persisters downregulated the expression of melanocytic differentiation proteins, which led to the development of a MITFlow/MDAlow tumor cell phenotype consistently detected in the distinct melanoma models studied. Transition into the MITFlow/MDAlow cell state was induced not only by 3pRNA but also by the synthetic RIG-I agonist poly(I:C). Among the dedifferentiated persisters, subpopulations evolved showing an upregulation also of the dedifferentiation markers NGFR or AXL. Treatment with Ruxolitinib largely blocked switching toward the MITFlow/MDAlow phenotype, suggesting an involvement of JAK-STAT signaling in RIG-I-induced cell state transition. In fact, 3pRNA treatment triggered phosphorylation of STAT1 and expression of IRF1 in melanoma cells. Activation of the JAK-STAT1-IRF1 pathway is also induced by interferons. IFN-γ has recently been described as a potent inducer of melanoma dedifferentiation, similar to TNF-α.15 22 39 Both inflammatory cytokines have been demonstrated to induce a Melan-Alow/NGFRhigh tumor-cell phenotype. Accordingly, we detected downregulation of MITF and its target genes in our melanoma models not only in response to 3pRNA but also to IFN-γ and TNF-α treatment. It remains to be determined whether mechanisms eliciting melanoma dedifferentiation on RIG-I signaling overlap with those triggered by IFN-γ or TNF-α. Moreover, it needs to be investigated whether other PRR ligands, currently tested in clinical trials, such as TLR agonists,42–44 have similar effects on the melanoma differentiation state.
Melanoma phenotype switching has repeatedly been associated with T cell and therapy resistance.15–17 22 45 Our own previous studies demonstrated that a subgroup of melanoma cells acquires resistance to MAPKi by transition into a dedifferentiated cell state that can confer cross-resistance to autologous tumor-reactive CD8 T cells.16 17 Unlike those MAPKi-resistant melanoma cells, 3pRNA-induced dedifferentiated persisters can still be efficiently recognized by autologous CD8 TILs which could be due to distinct transcriptional and epigenetic programs triggered in response to the specific treatments. According to its role in innate immunity, RIG-I signaling elicits the expression of specific immune molecules to ensure detection and killing by cytotoxic CD8 T cells. This includes several HLA-I APM components,2 which might reshape the presented antigen repertoire and boost recognition by autologous CD8 TILs.46 In fact, a recent study by Samuels and colleagues demonstrated that melanoma cells expressing subunits of the immunoproteasome display a distinct repertoire of HLA-I-bound tumor antigen peptides with stronger T cell-stimulatory capacity.47 Expression of immunoproteasome subunits is induced in response to RIG-I signaling2 and can be detected also in 3pRNA-induced persister cells. In this regard, 3pRNA-induced MITFlow/MDAlow persisters very much resemble IFN-γ-induced MITFlow/MDAlow melanoma cells, both showing JAK-STAT pathway activation linked to potent HLA-I APM upregulation. Tumor cell-intrinsic JAK-STAT signaling and HLA-I antigen presentation play a key role in response to immunotherapy.2 48 Notably, while TNF-α signaling also elicited melanoma dedifferentiation, it did not trigger JAK-STAT pathway activation and HLA-I APM upregulation, suggesting lack of immunogenicity enhancement for TNF-α-treated cells which should be addressed in future studies.
Hence, the outcome of treatment-induced dedifferentiation on anti-tumor T cell responses seems to be dependent on the agent that triggers melanoma dedifferentiation and the specific transcriptional program induced on treatment. In line with our findings, Kim et al recently demonstrated that melanoma biopsies from patients responding to ICB show a decrease in melanocytic marker expression.39 Based on this observation, the authors proposed melanoma dedifferentiation as an indicator for an initial response to anti-PD-1 therapy.39
Although data from our study and Kim et al challenge the concept of melanoma dedifferentiation as a general T cell and therapy resistance marker, we expect that the association still holds true for patients whose anti-tumor T cell responses heavily rely on a MDA-specific T cell repertoire, and for those receiving MDA-specific therapies, like adoptive transfer of MDA-specific T cells.
Overall, we think that our study is of strong clinical significance. First, our findings support ongoing clinical trials combining intratumoral injection of RIG-I agonists with anti-PD-1 blocking antibodies as a treatment option for unresectable melanomas (NCT03739138, NCT02828098). Second, our data suggest that treatment-induced melanoma dedifferentiation should not be considered as a general T cell and therapy resistance marker.
Acknowledgments
Cell sorting was performed at the Imaging Center Essen (IMCES), a Service Core Facility of the Faculty of Medicine of the University Duisburg-Essen, Germany.
Footnotes
Contributors: BT conceptualized the study, conducted experiments, acquired and analyzed data, and wrote the manuscript. FZ, SS and AB performed experiments, acquired and analyzed data, and reviewed the manuscript. JK and NS performed experiments, acquired, and analyzed data. SH provided software, conducted bioinformatics and statistical data analyses, and reviewed the manuscript. CC and GH provided critical reagents. AS and DS collected clinical samples and annotated clinical data. AP conceptualized and supervised the study, reviewed data, and wrote the manuscript. AP is responsible for the overall content as the guarantor.
Funding: AP was supported by grants of the Deutsche Krebshilfe (DKH, German Cancer Aid; Translational Oncology 70113455) and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - SFB1430— Project ID 424228829 and KFO337—Project ID 405344257 (PA 2376/1-1). DS was supported by grants of the Deutsche Krebshilfe (DKH, German Cancer Aid; Translational Oncology 70113455) and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) KFO337—Project ID 405344257 (SCHA 422/17-1). SH received funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) KFO337—Project ID 405344257 (HO 6389/2-1). GH was supported by grants of the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) of TRR259 and TRR237 and is member of Germany’s Excellence Strategy (EXC2151—390873048). NS was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - SFB1430—Project ID 424228829.
Competing interests: AP reports research grant support and provision of reagents from Bristol-Myers Squibb (BMS) and Merck Sharp & Dohme (MSD). AS reports personal fees from Novartis Adboard. GH is a co-founder of Rigontec GmbH. DS declares grants, personal fees, and/or nonfinancial support from BMS, Roche, Novartis, Regeneron, Sanofi, MSD, Amgen, 4SC, Merck-EMD, Array, Pierre-Fabre, Philogen, Incyte, and Pfizer.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Specimens from melanoma patients were collected after written informed consent. Studies on human material were approved by the institutional review board (18-8269-BO). Biological samples and related data were provided by the Westdeutsche Biobank Essen (WBE/SCABIO, University Hospital Essen, University of Duisburg-Essen, Essen, Germany; approval no. SCABIO_114715).
References
- 1.Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. 10.1016/j.cell.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Such L, Zhao F, Liu D, et al. Targeting the innate immunoreceptor RIG-I overcomes melanoma-intrinsic resistance to T cell immunotherapy. J Clin Invest 2020;130:4266–81. 10.1172/JCI131572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, Shklovskaya E, Lim SY, et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat Commun 2020;11:1–12. 10.1038/s41467-020-15726-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Middleton MR, Hoeller C, Michielin O, et al. Intratumoural immunotherapies for unresectable and metastatic melanoma: current status and future perspectives. Br J Cancer 2020;123:885–97. 10.1038/s41416-020-0994-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melero I, Castanon E, Alvarez M, et al. Intratumoural administration and tumour tissue targeting of cancer immunotherapies. Nat Rev Clin Oncol 2021;18:558–76. 10.1038/s41571-021-00507-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bommareddy PK, Silk AW, Kaufman HL. Intratumoral approaches for the treatment of melanoma. Cancer J 2017;23:40–7. 10.1097/PPO.0000000000000234 [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, Ellegast J, Kim S, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science 2006;314:994–7. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 8.Iurescia S, Fioretti D, Rinaldi M. The innate immune signalling pathways: turning RIG-I sensor activation against cancer. Cancers 2020;12:3158–26. 10.3390/cancers12113158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalbasi A, Tariveranmoshabad M, Hakimi K, et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med 2020;12:152. 10.1126/scitranslmed.abb0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torrejon DY, Abril-Rodriguez G, Champhekar AS, et al. Overcoming genetically based resistance mechanisms to PD-1 blockade. Cancer Discov 2020;10:1140–57. 10.1158/2159-8290.CD-19-1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidegger S, Wintges A, Stritzke F, et al. RIG-I activation is critical for responsiveness to checkpoint blockade. Sci Immunol 2019;4:8943. 10.1126/sciimmunol.aau8943 [DOI] [PubMed] [Google Scholar]
- 12.Aznar MA, Planelles L, Perez-Olivares M, et al. Immunotherapeutic effects of intratumoral nanoplexed poly I:C. J Immunother Cancer 2019;7:1–16. 10.1186/s40425-019-0568-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boumahdi S, de Sauvage FJ. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov 2020;19:39–56. 10.1038/s41573-019-0044-1 [DOI] [PubMed] [Google Scholar]
- 14.Rambow F, Marine J-C, Goding CR. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev 2019;33:1295–318. 10.1101/gad.329771.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landsberg J, Kohlmeyer J, Renn M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012;490:412–6. 10.1038/nature11538 [DOI] [PubMed] [Google Scholar]
- 16.Pieper N, Zaremba A, Leonardelli S. Evolution of melanoma cross-resistance to CD8+ T cells and MAPK inhibition in the course of BRAFi treatment. Oncoimmunology 2018;7:e1450127. 10.1080/2162402X.2018.1450127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbers FN, Thier B, Stupia S, et al. Melanoma differentiation trajectories determine sensitivity toward pre-existing CD8+ tumor-infiltrating lymphocytes. J Invest Dermatol 2021;141:2480–9. 10.1016/j.jid.2021.03.013 [DOI] [PubMed] [Google Scholar]
- 18.Restivo G, Diener J, Cheng PF, et al. Low neurotrophin receptor CD271 regulates phenotype switching in melanoma. Nat Commun 2017;8:1988. 10.1038/s41467-017-01573-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rambow F, Rogiers A, Marin-Bejar O, et al. Toward minimal residual Disease-Directed therapy in melanoma. Cell 2018;174:843–55. 10.1016/j.cell.2018.06.025 [DOI] [PubMed] [Google Scholar]
- 20.Konieczkowski DJ, Johannessen CM, Abudayyeh O, et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov 2014;4:816–27. 10.1158/2159-8290.CD-13-0424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller J, Krijgsman O, Tsoi J, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014;5:5712. 10.1038/ncomms6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta A, Kim YJ, Robert L, et al. Immunotherapy resistance by inflammation-induced dedifferentiation. Cancer Discov 2018;8:935–43. 10.1158/2159-8290.CD-17-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugo W, Shi H, Sun L, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 2015;162:1271–85. 10.1016/j.cell.2015.07.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. 10.1016/j.cell.2016.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao F, Sucker A, Horn S, et al. Melanoma lesions independently acquire T-cell resistance during metastatic latency. Cancer Res 2016;76:4347–58. 10.1158/0008-5472.CAN-16-0008 [DOI] [PubMed] [Google Scholar]
- 26.Sucker A, Zhao F, Pieper N, et al. Acquired IFNγ resistance impairs anti-tumor immunity and gives rise to T-cell-resistant melanoma lesions. Nat Commun 2017;8:15440. 10.1038/ncomms15440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsoi J, Robert L, Paraiso K, et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 2018;33:890–904. 10.1016/j.ccell.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gide TN, Quek C, Menzies AM, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and Anti-PD-1/Anti-CTLA-4 combined therapy. Cancer Cell 2019;35:238–55. 10.1016/j.ccell.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 29.Liu D, Schilling B, Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916–27. 10.1038/s41591-019-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 31.Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturm G, Finotello F, Petitprez F, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019;35:i436–45. 10.1093/bioinformatics/btz363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besch R, Poeck H, Hohenauer T, et al. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. J Clin Invest 2009;119:2399–411. 10.1172/JCI37155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kübler K, Gehrke N, Riemann S, et al. Targeted activation of RNA helicase retinoic acid-inducible gene-I induces proimmunogenic apoptosis of human ovarian cancer cells. Cancer Res 2010;70:5293–304. 10.1158/0008-5472.CAN-10-0825 [DOI] [PubMed] [Google Scholar]
- 36.Duewell P, Steger A, Lohr H, et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ 2014;21:1825–37. 10.1038/cdd.2014.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carreira S, Goodall J, Denat L, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 2006;20:3426–39. 10.1101/gad.406406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boshuizen J, Vredevoogd DW, Krijgsman O, et al. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat Commun 2020;11:1–13. 10.1038/s41467-020-17739-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim YJ, Sheu KM, Tsoi J, et al. Melanoma dedifferentiation induced by IFN-γ epigenetic remodeling in response to anti–PD-1 therapy. J Clin Invest 2021;131. 10.1172/JCI145859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elion DL, Jacobson ME, Hicks DJ, et al. Therapeutically active RIG-I agonist induces immunogenic tumor cell killing in breast cancers. Cancer Res 2018;78:6183–95. 10.1158/0008-5472.CAN-18-0730 [DOI] [PubMed] [Google Scholar]
- 41.Jiang X, Muthusamy V, Fedorova O, et al. Intratumoral delivery of RIG-I agonist SLR14 induces robust antitumor responses. J Exp Med 2019;216:2854–68. 10.1084/jem.20190801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribas A, Medina T, Kummar S, et al. Sd-101 in combination with pembrolizumab in advanced melanoma: results of a phase Ib, multicenter study. Cancer Discov 2018;8:1250–7. 10.1158/2159-8290.CD-18-0280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribas A, Medina T, Kirkwood JM, et al. Overcoming PD-1 blockade resistance with CpG-A Toll-like receptor 9 agonist Vidutolimod in patients with metastatic melanoma. Cancer Discov 2021;11:candisc.0425.2021 10.1158/2159-8290.CD-21-0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Márquez-Rodas I, Longo F, Rodriguez-Ruiz ME, et al. Intratumoral nanoplexed poly I:C BO-112 in combination with systemic anti-PD-1 for patients with anti-PD-1-refractory tumors. Sci Transl Med 2020;12:391. 10.1126/scitranslmed.abb0391 [DOI] [PubMed] [Google Scholar]
- 45.Massi D, Mihic-Probst D, Schadendorf D, et al. Dedifferentiated melanomas: morpho-phenotypic profile, genetic reprogramming and clinical implications. Cancer Treat Rev 2020;88:102060. 10.1016/j.ctrv.2020.102060 [DOI] [PubMed] [Google Scholar]
- 46.Olsson N, Heberling ML, Zhang L, et al. An integrated genomic, proteomic, and immunopeptidomic approach to discover treatment-induced neoantigens. Front Immunol 2021;12:1–17. 10.3389/fimmu.2021.662443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalaora S, Lee JS, Barnea E, et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat Commun 2020;11:1–12. 10.1038/s41467-020-14639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paschen A, Melero I, Ribas A. Central role of the antigen-presentation and interferon-γ pathways in resistance to immune checkpoint blockade. Annu Rev Cancer Biol 2022;6:85–102. 10.1146/annurev-cancerbio-070220-111016 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003863supp001.pdf (5.1MB, pdf)
jitc-2021-003863supp002.xlsx (265.3KB, xlsx)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.