FIG. 5.
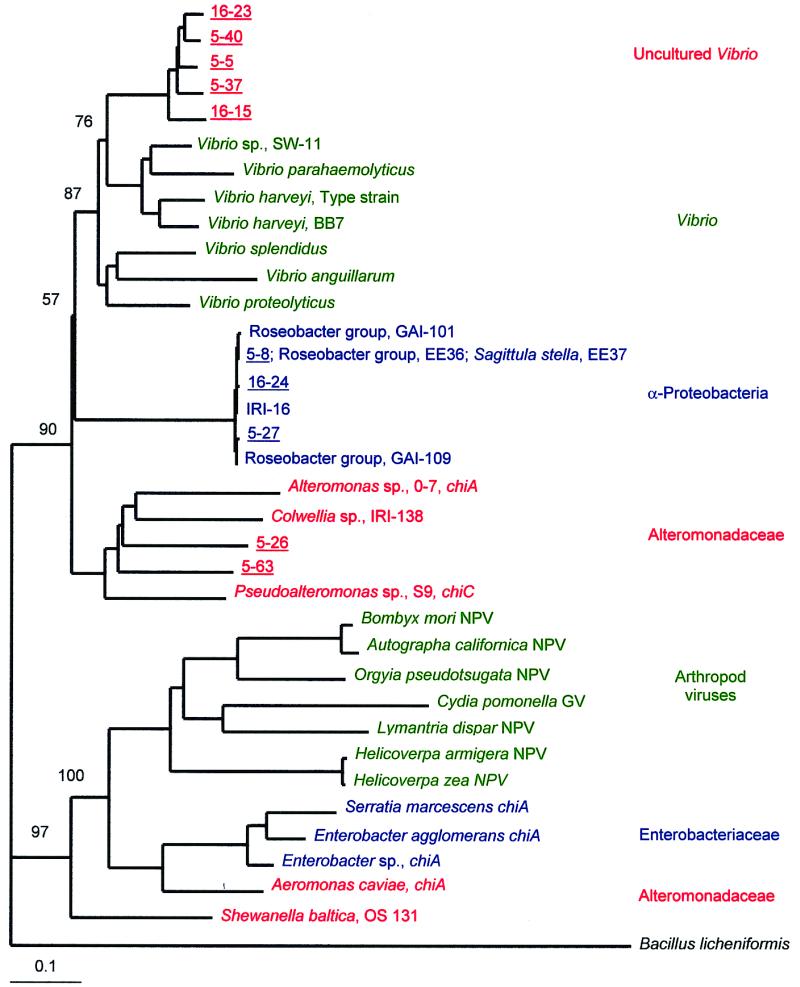
Additive phylogenetic tree of group I chitinase genes from cultured and uncultured bacteria and viruses of arthropods. The neighbor-joining analysis used a chitinase gene from B. licheniformis (GenBank accession no. U71214) as the outgroup. α-Proteobacteria (15, 16) and γ-proteobacteria were isolated from coastal and estuarine environments. Shewanella baltica was isolated from the Baltic Sea (42). The nucleotide sequences of the chitinase genes of reference strains of Pseudoalteromonas sp. (38), Enterobacter sp. (33), B. mori NPV (23), Autographa californica NPV (4), Orgyia pseudotsugata NPV (1), Helicoverpa zea NPV (26), Helicoverpa armigera NPV (GenBank accession no. AF114795), Lymantria dispar NPV (25), and Cydia pomonella granulovirus (GV) (24) were obtained from GenBank. Bootstrap values are indicated at the nodes separating the major groups. Genes from uncultured bacteria are underlined and are designated by a station and clone number (e.g., 5-40). The scale bar indicates the amount of genetic change measured as the number of nucleotide substitutions per site.
