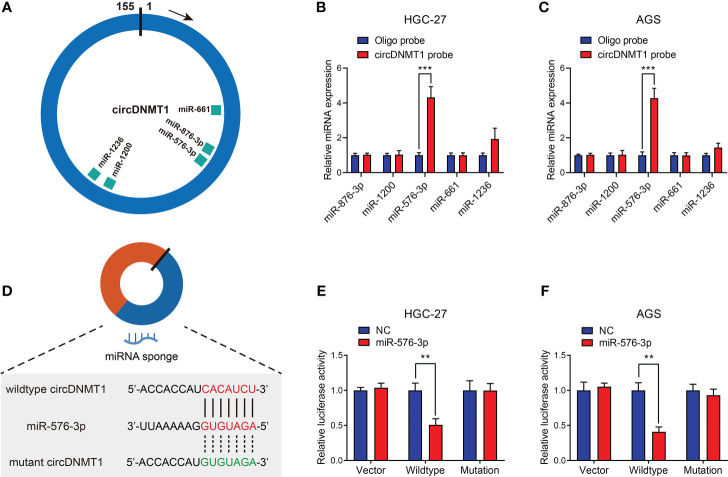
Figure 4.
miR-576-3p is directly sponged by circDNMT1 in GC. (A) The schematic illustration of miRNAs that can be sponged by circDNMT1 predicted by Circinteractome. (B, C) The qRT-PCR analysis to show the miRNA levels that were pulled down by NC probe or circDNMT1 probe in HGC-27 (B) and AGS (C) cells. (D) The schematic illustration of wildtype (red) and mutant (green) sequences of binding sites of circDNMT1 and miR-576-3p. (E, F) Luciferase reporter assay to show the relative luciferase activities in HGC-27 (E) and AGS (F) cells that were cotransfected with empty luciferase reporter plasmids (vector) or plasmids inserted with wildtype or mutant sequences and NC and miR-567-3p mimics. **P < 0.01, ***P < 0.001.