Figure 2. Mass-Spectrometry Techniques for Protein Conformational Stability Measurements.
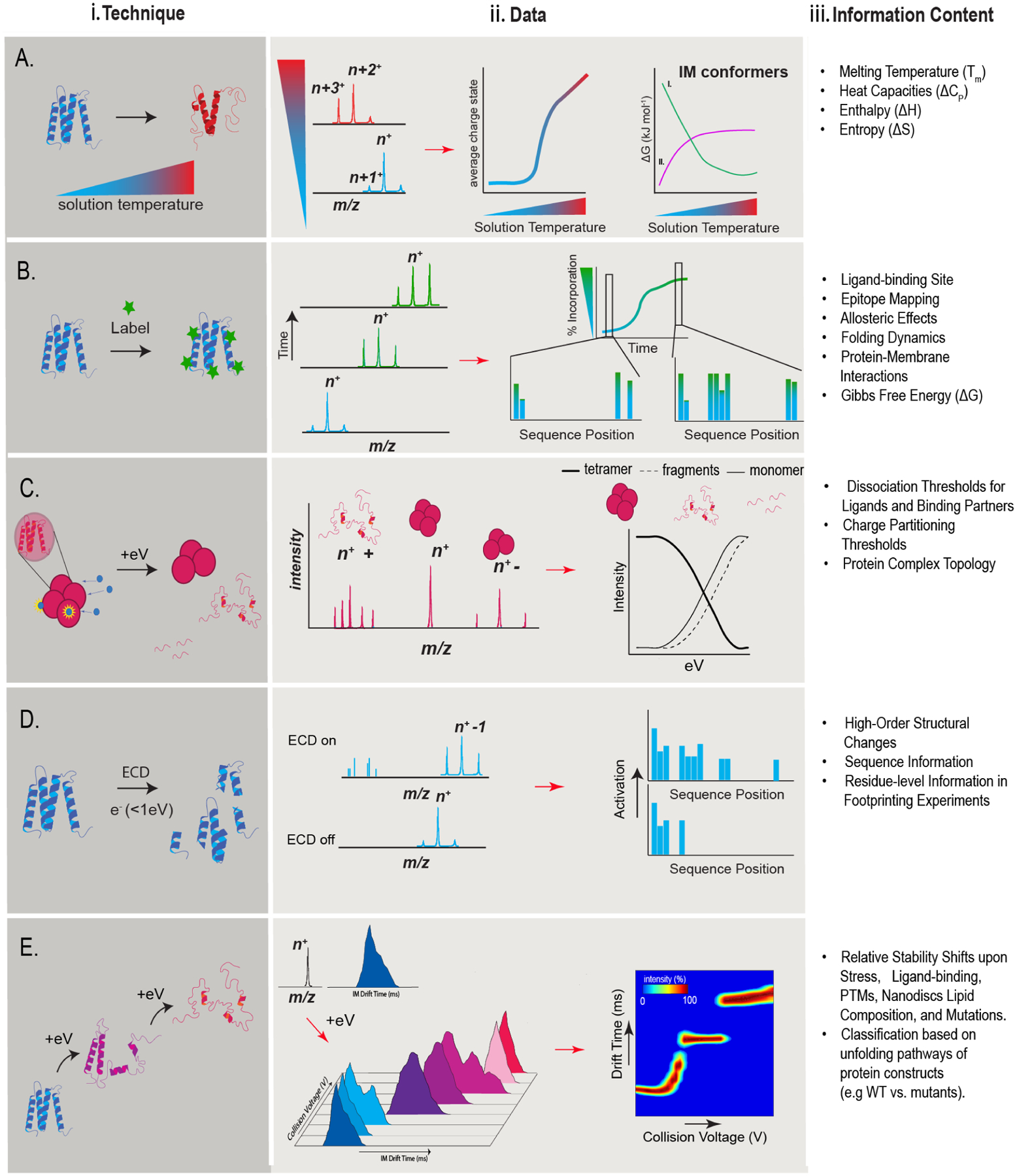
For each technique described, there is a corresponding summary figure, ii) data and iii) information content column. A) Variable Temperature. A protein experiences a temperature gradient as it is introduced into the mass spectrometer. A shift in CSD indicates conformational changes. By monitoring IM conformers, insights into thermodynamic values can be made. B) Footprinting uses selective or non-selective reagents (green stars) to monitor changes to the solvent-accessible regions of a protein. C) Electron Capture Dissociation is a fragmentation technique where proteins are exposed to low energy electrons, which are captured and produce backbone fragmentation. ECD fragmentation patterns can changed based on the precursor conformation and charge state. D) Collision Induce Dissociation allows the evaluation of protein complex subunit stoichiometry and composition. By monitoring protein ejection from protein complex precursors, dissociation thresholds can be determined and related to the stabilities of subunits and interfaces. E) Collision Induced Unfolding is an IM-MS technique where a protein conformation is monitored as its internal energy is increased using collisional activation. A shift in voltage required for eliciting the unfolding transitions observed is indicative of stability shifts when comparing between fingerprints of different states (e.g. apo vs holo).
