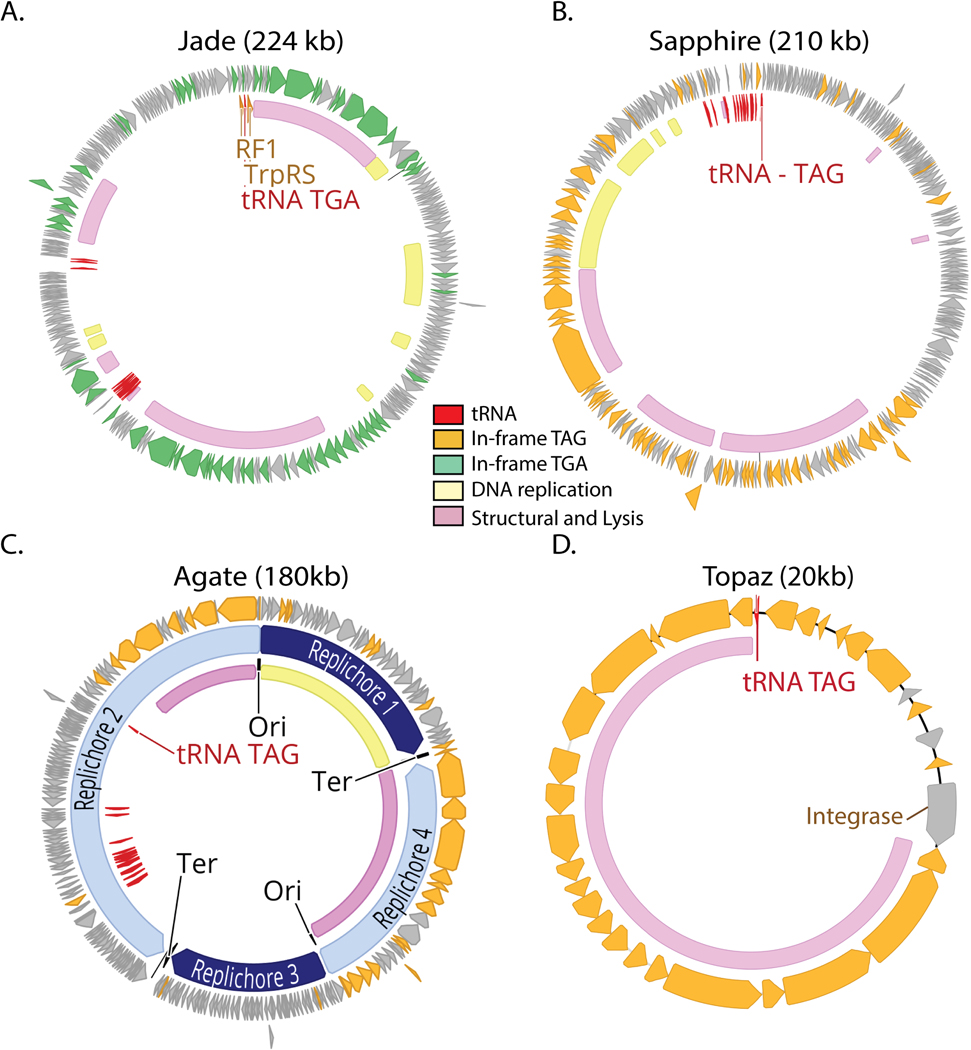
Extended Data Fig. 4. Genomic maps of Jade, Sapphire, Agate and Topaz phages.
A-D. TGA recoded genomes (A) contain genes with in-frame TGA codons (green) while TAG recoded genomes (B-D) have genes with in-frame TAG codons (orange). Suppressor tRNAs (tRNA TGA or tRNA TAG, red) are predicted to suppress translation termination at TGA and TAG stop codons, respectively. Regions of the genome encoding structural and lysis genes (pink) coincide with high use of alternative code. Contrastingly, genes involved in DNA replication (yellow) are variably encoded in alternative code. Genomes with a GC skew patterns indicative of bidirectional replication and clear origins and termini (C) have unique replichores marked in alternating shades of blue. Genomes with GC skew patterns most consistent with unidirectional replication (A-B,D) have no replication-related annotation. In some cases, unique or interesting genes have been noted with text. Clade representatives: Jade = JS_HF2_S141_scaffold_159238, Sapphire= SRR1747018_scaffold_13, Agate = Cattle_ERR2019359_scaffold_1067472, Topaz = pig_ID_1851_F40_2_B1_scaffold_1589