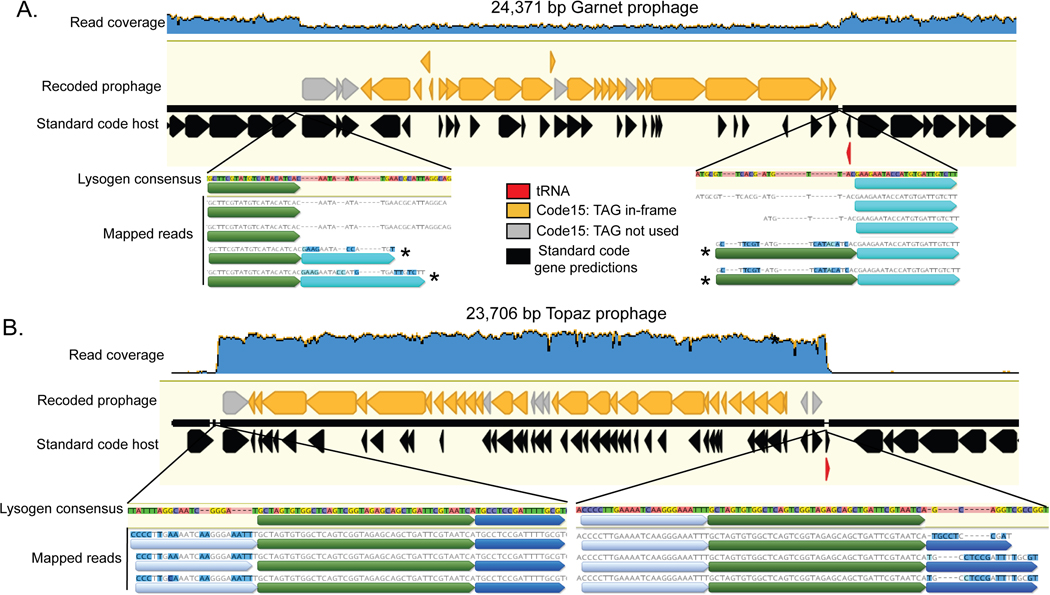
Extended Data Fig. 7. Read mapping to Garnet and Topaz lysogens.
A. Reads were mapped against a manually curated Garnet lysogen. Read coverage for the Prevotella DNA is ∼2x higher than the read coverage of the Garnet prophage, indicating that the bacterial population in this sample is incompletely lysogenized. Supporting this conclusion are paired reads that span the prophage (not shown), as well as some individual reads which show imperfect mapping to the lysogen consensus sequence (marked with asterisk), which represent the contiguous bacterial sequence. Identical sequence blocks are indicated with color. B. Reads were mapped against a manually curated Topaz lysogen. Read coverage for the integrated Topaz phage genome is ∼50x higher than the neighboring Oscillospiraceae sequence. This indicates that the phage is actively replicating in this sample. Supporting this conclusion are paired reads that span the length of the prophage (not shown), as well as individual reads which show imperfect mapping to the lysogen consensus sequence at the 5’ end of the prophage (light blue) and the 3’ end of the prophage (dark blue). The reads correspond to circularized sequences. Identical sequence blocks are indicated with color.