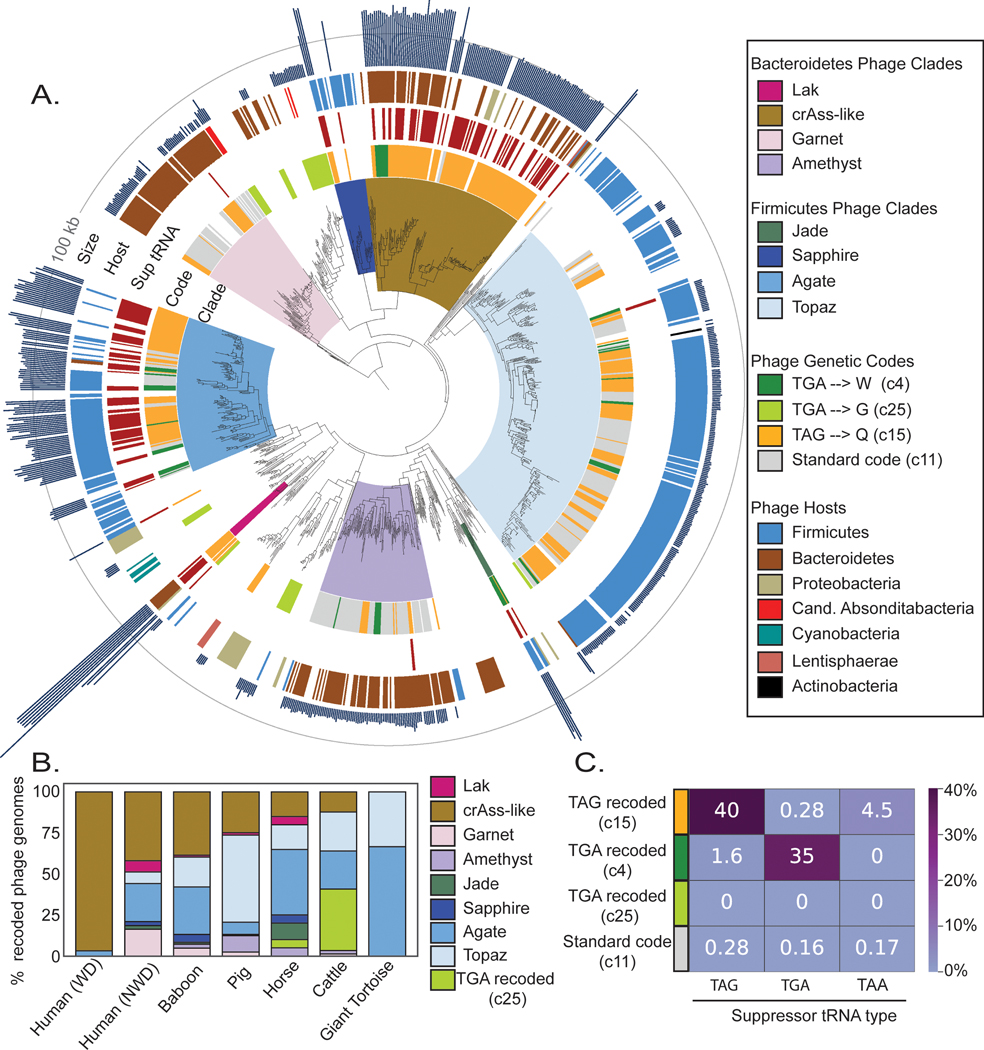
Fig. 2: Phylogeny of recoded phages and suppressor tRNA usage.
A. The phylogeny of recoded phages was reconstructed using large terminase sequences from a dereplicated set of complete or near-complete (>=90%) recoded phages (n=444) and their close standard code relatives (n =258), as well as related proteins from Refseq r205 (n=410). Terminase sequences from eukaryotic herpesviruses (n=8) were used to root the tree. The inner to outer ring shows phage clade (>= 95% bootstrap support), genetic code for phages from this study, suppressor tRNA presence, host phylum as predicted by taxonomic profiling and CRISPR spacer matches, and genome size with a grey line at 100 kilobases (kb) for scale. Genetic code, suppressor tRNA presence, and genome size were not included for Refseq proteins since some proteins were derived from prophages and/or incomplete phage genomes.
B. Distribution of recoded phages by clade across the 7 types of gut microbiomes evaluated in this study. WD = westernized diet, NWD = non-westernized diet.
C. Heatmap of the percent of genomes of each genetic code that have tRNAs predicted to suppress translation termination at the TAG, TGA, or TAA stop codons.