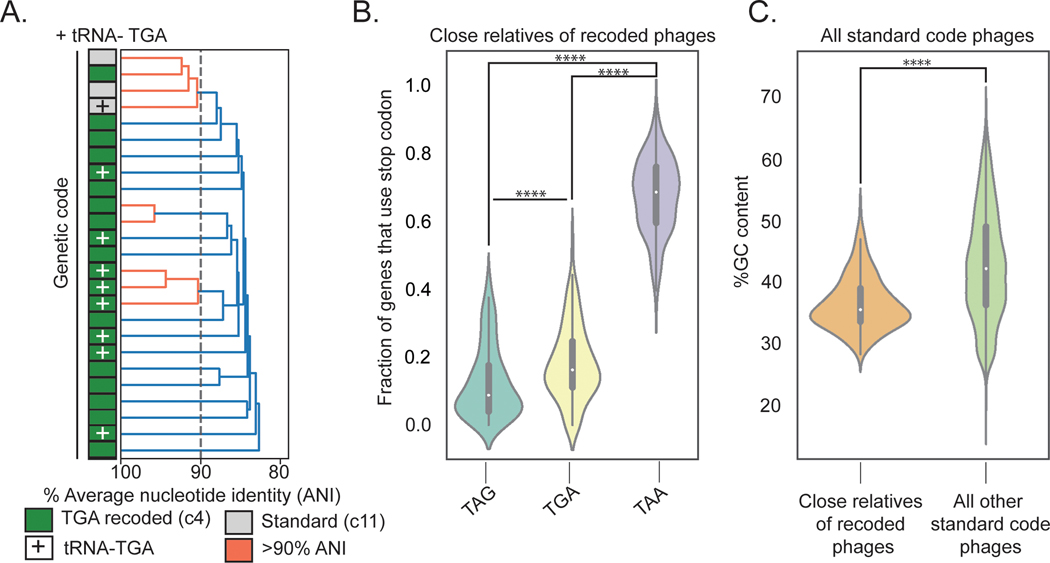
Fig. 3: Evolutionary relationships among phages according to genetic code.
A. Dendrogram of average nucleotide identity (ANI) across a set of Agate phage genomes. Standard code (grey) and TGA recoded (green) phages share >80% ANI, and one cluster of >90% ANI genomes (orange) has both standard and TGA recoded genomes, indicating an extremely close evolutionary relationship.
B. Close relatives of recoded phages (n = 260 biologically independent phage genomes) use the TAA stop codon codon at a higher rate than the TAG and TGA stop codons (TAG vs. TAA: Z = −19.71, p = 1.63e-86, TGA vs. TAA: Z = −19.65, p = 5.98e-86, two-sided Wilcoxon Rank-Sum Test). The TAG stop codon is depleted relative to TGA (Z = −6.43, p = 1.24e-10, two-sided Wilcoxon Rank-Sum Test) in these phages. TAG frequencies: Minima = 0.0, Maxima = 0.38, Median = 0.09, IQR = 0.14, Q1 = 0.04 Q3 = 0.18. TGA frequencies: Minima = 0.0, Maxima = 0.44, Median = 0.16, IQR = 0.13, Q1 = 0.11, Q3 = 0.25. TAA frequencies: Minima = 0.40, Maxima = 0.94, Median = 0.68, IQR = 0.16, Q1 = 0.59, Q3 = 0.76.
C. Close relatives of alternatively coded phages have a lower mean GC content relative to all other standard code phages (Z = −12.59, p = 2.33e-36, two-sided Wilcoxon Rank-Sum Test). Close relatives: n = 260 biologically independent phage genomes, Minima = 27.97, Maxima = 45.73, Median = 35.10, IQR = 5.30, Q1 = 33.27, Q3 = 38.57. Unrelated standard code phages: n = 8689 biologically independent phage genomes, Minima = 19.60, Maxima = 67.07, Median = 41.827, IQR = 12.67, Q1 = 35.95, Q3 = 48.62.
**** p ≤ 0.0001, two-sided Wilcoxon Rank-Sum test.