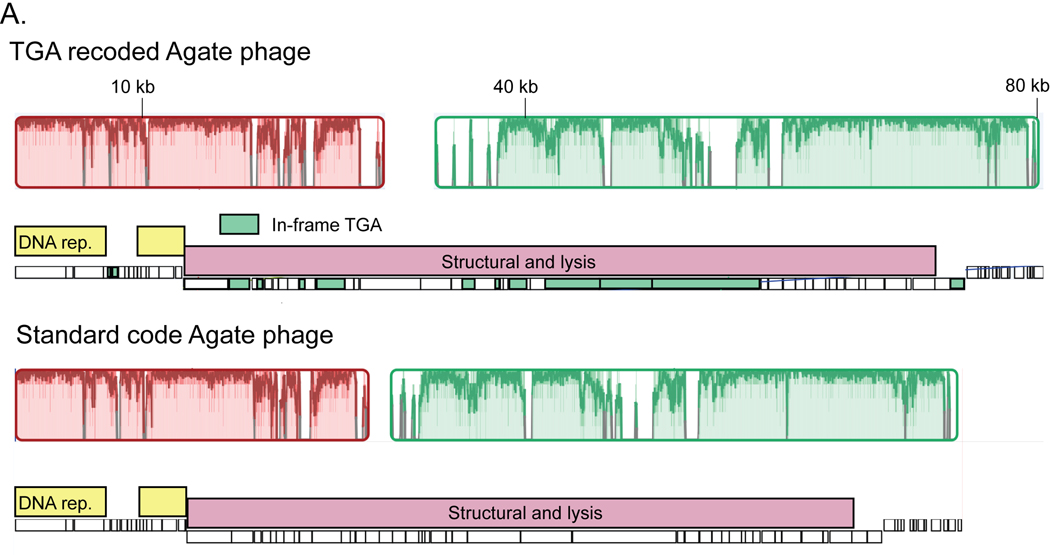
Extended Data Fig. 3. Evolution of alternative coding.
A. Global alignment of an 80 kilobase (kb) partial TGA recoded Agate genome (Cattle_ERR2019405_scaffold_1063) and a close standard code relative (pig_ID_3053_F60_scaffold_12). Homologous collinear sequences are shown with colored blocks (red and green here), where color corresponds to nucleotide alignment between the two genomes and lack of color represents lack of alignment. Genome structure for each phage is shown under the alignment graph, with DNA replication machinery represented as yellow bars and structural and lysis genes with pink bars. TGA stop codons have predominantly arisen in structural and lysis genes (individual recoded genes below in green).