Abstract
Background
Excessive inflammation is pathogenic in the pneumonitis associated with severe COVID-19. Neutrophils are among the most abundantly present leukocytes in the inflammatory infiltrates and may form neutrophil extracellular traps (NETs) under the local influence of cytokines. NETs constitute a defense mechanism against bacteria, but have also been shown to mediate tissue damage in a number of diseases.
Research Question
Could NETs and their tissue-damaging properties inherent to neutrophil-associated functions play a role in the respiratory failure seen in patients with severe COVID-19, and how does this relate to the SARS-CoV-2 viral loads, IL-8 (CXCL8) chemokine expression, and cytotoxic T-lymphocyte infiltrates?
Study Design and Methods
Sixteen lung biopsy samples obtained immediately after death were analyzed methodically as exploratory and validation cohorts. NETs were analyzed quantitatively by multiplexed immunofluorescence and were correlated with local levels of IL-8 messenger RNA (mRNA) and the density of CD8+ T-cell infiltration. SARS-CoV-2 presence in tissue was quantified by reverse-transcriptase polymerase chain reaction and immunohistochemistry analysis.
Results
NETs were found in the lung interstitium and surrounding the bronchiolar epithelium with interindividual and spatial heterogeneity. NET density did not correlate with SARS-CoV-2 tissue viral load. NETs were associated with local IL-8 mRNA levels. NETs were also detected in pulmonary thrombi and in only one of eight liver tissues. NET focal presence correlated negatively with CD8+ T-cell infiltration in the lungs.
Interpretation
Abundant neutrophils undergoing NETosis are found in the lungs of patients with fatal COVID-19, but no correlation was found with viral loads. The strong association between NETs and IL-8 points to this chemokine as a potentially causative factor. The function of cytotoxic T-lymphocytes in the immune responses against SARS-CoV-2 may be interfered with by the presence of NETs.
Key Words: COVID-19, lungs, neutrophil extracellular traps, neutrophils, SARS-CoV-2
Abbreviations: FFPE, formalin-fixed, paraffin-embedded; mRNA, messenger RNA; NET, neutrophil extracellular trap
Take-home Points.
Study Question: Are neutrophils and neutrophil extracellular traps (NETs) involved in lung tissue damage in severe COVID-19?
Results: We detected NETs in all 16 lung biopsy samples obtained immediately after death from patients with fatal COVID-19. Moreover, we observed heterogenous spatial distribution of the NETs, whose abundance was unrelated to viral load as assessed by reverse-transcriptase polymerase chain reaction and immunohistochemistry. In contrast, NET density correlated with local expression of the IL-8 chemokine at the messenger RNA and protein levels. This chemokine is known both to chemoattract neutrophils and to cause NET extrusion. In this collection of tissue samples obtained immediately after death, areas rich in NETs tended to exclude the presence of CD8 T lymphocytes.
Interpretation: Our study provides unequivocal evidence for the abundant presence of NETs in all the patients with fatal COVID-19 studied. NETs are likely to be pathogenic in COVID-19 because they induce tissue damage and thrombosis, and therefore it would be logical to interfere with neutrophil infiltration or the NETosis pathway by means pharmacologic interventions. IL-8 is known to be associated with NETosis induction acting on the CXCR1 and CXCR2 receptors and is associated spatially with NETs in lungs of patients with fatal COVID-19. Anti-IL-8 monoclonal antibodies are being tested in several cancer clinical trials, and CXCR1 and CXCR2 pharmacologic inhibitors are also available. Drugs currently under development inhibiting the peptidyl arginine deiminase 4 enzyme to inhibit NETosis could also be repurposed to this end.
COVID-19 caused by SARS-CoV-2 infection is fatal in 1% to 3% of patients, although this has been much mitigated with the availability of prophylactic vaccines.1, 2, 3 SARS-CoV-2-induced pneumonia likely begins with viral replication inducing direct virus-mediated tissue damage. Leukocytes are then recruited, producing a local and systemic inflammatory response that can persist even after viral clearance.4 The extent of tissue damage seems to be related more to excessive inflammation than to direct virus effects.5 Lung tissue from patients with fatal COVID-19 often reveals a spectrum of diffuse alveolar damage associated with excess vascular permeability leading to microthrombi formation and varying degrees of inflammation and types of cellular infiltrates.6, 7, 8, 9 An excess of circulating myeloid progenitor cells, immature monocytes, and neutrophils is associated with COVID-19 disease severity.10 , 11 These circulating cells can be the source of excessive amounts of inflammatory mediators that promote vascular permeability and ultimately cause organ damage.4
Neutrophils are found in the lungs of patients with fatal COVID-19.12 Although their role in COVID-19 pathogenesis remains unclear, neutrophil-associated functions, such as the production of reactive oxygen species, proteases, inflammatory mediators, and cytotoxicity, potentially are harmful mechanisms, at least in the lungs. Neutrophil extracellular traps (NETs) are the result of a form of neutrophil death, called NETosis.13, 14, 15 Neutrophils extrude their nuclear DNA coated with lysosomal and cytoplasmatic polypeptides as a defense mechanism to entrap and kill prokaryotic microbes. NETs are also involved in a wide variety of diseases, including chronic lung diseases.16 In bronchiectasis, NETs correlate with worse outcome.17
NETs are characterized by nucleosomes of double-stranded DNA bound to citrullinated histone 3, neutrophil elastase, and myeloperoxidase. These proteins are adsorbed onto the DNA web complexes. Many microbial components and inflammatory mediators have been shown to induce NETosis. ELR+ CXCL chemokines acting on CXCR1 and CXCR2, such as IL-8 (CXCL8), are among the most effective NET promoters.13
SARS-CoV-2 has also been shown to elicit NETosis directly on human neutrophils.18 In patients with COVID-19, NETs have been detected in the circulation and lung tissue, including the vascular and airway compartment.9 , 18, 19, 20, 21 Therefore, NETs have been proposed as being pathogenic in COVID-19.
In this study, we investigated a series of 16 consecutive lung and liver postmortem biopsy samples obtained immediately after death from patients with COVID-19. A multiplexed immunofluorescence assay to detect and quantify NETs was applied to study the spatial distribution of NETs and its correlation with tissue SARS-CoV-2 viral load, clinical features, and the presence of local IL-8 and CD8+ T cells.
Study Design and Methods
Ethics Statement
This study was approved by the ethics committee of the University of Navarra, Spain (Identifier: 2020.192). Tissue samples were obtained with consent from a first-degree relative, following a protocol approved by the ethics committee of the University of Navarra (protocol no.: 2020.192p). Tissues were stored at the Clínica Universidad de Navarra according to Human Tissue Authority regulations.
Patient Cohorts
Postmortem lung tissues from 16 patients with a diagnosis of severe COVID-19 were collected and processed as described.6 Liver tissue was available from eight patients. Patients were divided into an exploratory cohort (n = 6) and a validation cohort (n = 10) based on the date of biopsy. Only patients with negative bacterial culture results at or before the time of death were included in this study. The demographic and clinical characteristics of the patients with COVID-19 and control participants are provided in e-Table 1. Tissues from six patients with ARDS unrelated to COVID-19 were included as a control group. The underlying causes of ARDS and patient demographics are detailed in e-Table 1. Pathologic features are shown in e-Table 2. Three histologically normal lung tissues collected from patients undergoing lung cancer surgery and three bacterial pneumonias unrelated to COVID-19 obtained from autopsy samples were also included as control tissues.
RNA Extraction and Quantitative Reverse-Transcriptase Polymerase Chain Reaction
Total RNA was extracted from two 5-μm formalin-fixed, paraffin-embedded (FFPE) tissue sections using the miRNeasy FFPE Kit (Qiagen). SARS-CoV-2 envelope (E) gene and nucleocapsid (N) gene were quantified using a commercial kit (SARS-CoV-2 Real Time PCR Kit; Vircell). Samples were classified as showing positive results for SARS-CoV-2 when at least one of the two SARS-CoV-2 genes was detected with a Ct value of < 35.
Immunohistochemistry Analysis for SARS-CoV-2 Nucleocapsid Protein
Staining was conducted with the Bond Polymer Refine Detection kit (Leica Biosystems), a rabbit anti-SARS-CoV-2 nucleocapsid antibody (clone, 001; dilution, 1:5000; Sinobiological), and counterstained with hematoxylin using a Leica BOND-RXm (Leica Biosystems). Whole-slide image analysis and virus quantification were performed with the open-source digital pathology software QuPath version 0.2.3 (University of Edinburgh). The number of positive cells was calculated as a percentage of the total number of cells.
Quantitative In Situ IL-8 Messenger RNA Measurement
In situ detection of IL-8 messenger RNA (mRNA) transcripts in FFPE tissue sections was carried out using the RNAscope assay (Advanced Cell Diagnostics) coupled to quantitative immunofluorescence. Whole-slide image analysis and IL-8 mRNA quantification was performed using ImageJ version 1.52c software (National Institutes of Health). Briefly, the fluorescence signal level of IL-8 mRNA was measured in the cellular compartment given by an expansion of each detected nucleus that creates an approximation of the full cell area.
Immunohistochemistry Analysis for IL-8 Protein
Detection of IL-8 protein in FFPE tissue sections was conducted with EnVision FLEX Target Retrieval Solution, high pH (Dako), a monoclonal mouse anti-IL8 antibody (clone, 807; dilution, 1:100; Abcam) and counterstained with hematoxylin. Whole-slide image analysis was performed using open-source software QuPath version 0.2.3 and is detailed in e-Figure 1.
Multiplexed Immunofluorescence Assays
Two previously validated and standardized multiplexed immunofluorescence protocols were used (optimization in e-Fig 2).22 , 23 Each multiplexed panel allowed for the simultaneous visualization and quantification of several cellular markers in a single FFPE tissue section. The NETosis multiplexed panel included a mouse monoclonal anti-CD15 antibody (clone, Carb-3; dilution, 1:100; Agilent), a rabbit monoclonal antimyeloperoxidase antibody (clone, E1E7I; isotype, IgG; dilution, 1:2,000; Cell Signaling Technology), a rabbit polyclonal anti-histone H3 antibody (citrulline R2+R8+R17, H3Cit; dilution, 1:100; Abcam), a mouse monoclonal anti-CD8 antibody (clone, C8/144B; isotype IgG1, kappa, ready to use; Agilent), a polyclonal rabbit anticytokeratin antibody (wide spectrum screening; dilution, 1:100; Agilent), and DAPI. The myeloid and lymphoid cell multiplexed panel included a mouse monoclonal anti-CD68 antibody (clone, PG-M1, ready to use; Agilent), a rabbit polyclonal anti-CD3 antibody (IgG, ready to use; Agilent), a mouse monoclonal anti-CD8 antibody (clone, C8/144B, ready to use; Agilent), a mouse monoclonal anti-CD20 antibody (IgG2α; clone, L26, ready to use; Agilent), a mouse monoclonal anti-CD66b antibody (clone, 80H3; dilution, 1:200; LS-Bio), a mouse monoclonal anticytokeratin antibody (clone, AE1/AE3, ready to use; Leica Biosystems), and DAPI. Briefly, FFPE tissue sections were subjected to sequential rounds of antibody staining. Secondary antibody incubation was carried out with a mix of horseradish peroxidase conjugated antimouse and antirabbit secondary antibodies (Akoya Bioscience) and tyramide signal amplification visualization with fluorophores Opal 520, 540, 570, 620, and 650 (Akoya Bioscience), as described previously.22 , 23 An additional double-staining fluorescence panel included granzyme B (IgG2α; clone, 4E6; dilution, 1:50; Abcam) to map cytolytic function on CD8+ T cells.
Tissue Imaging and Image Analysis
Multiplexed immunofluorescence slides were scanned on a Vectra-Polaris Automated Quantitative Pathology Imaging System (Akoya Biosciences). Spectral unmixing was performed using inForm software version 2.4.8 (Akoya Biosciences), as described.22 , 23 Image analysis was performed using the open-source software QuPath version 0.2.3 and Fiji/Image J. A grid of tiles (1 mm2) covering the whole-tissue section was applied in each image to quantify NETs and CD8+ T cells. Both whole-tissue analysis and tile analysis were performed to calculate the density of NETs (percentage area) and the percentage of CD8+ T cells. The immune infiltrate evaluated with the myeloid and lymphoid cell multiplexed panel was quantified in 20 randomly selected tiles. For control tissues, whole-tissue analysis and tile analysis were also performed. Because of specimen size, normal lung tissues and bacterial pneumonias comprised between two and four tiles.
NETosis processes were detected and measured based on the costaining of CD15, myeloperoxidase, and H3Cit by a plugin in Image J software as described.22 Briefly, in each image, Otsu’s thresholding algorithm was applied to establish single threshold levels of detection for each individual marker. This automated thresholding method allowed the detections acquired at different exposure times to be compared. The area of NETosis then was measured. Whole-tissue area and tile area were used to normalize the NETosis area and to determine the percentage area of NETs.
Myeloid and lymphoid cells were phenotyped as described.23 Briefly, cells were segmented based on nuclear detection using the StarDist 2D algorithm. A random trees algorithm classifier was trained for each cell marker. Cells then were subclassified as CD68+, CD3+, CD8+, CD66b+, and CD20+ cells. CD4+ T cells were defined as CD3+ CD8–. In addition, the expression levels of granzyme B were measured in CD8+ T cells. Cells negative for these markers were defined as other cell types. Measurements were calculated as percentage of the total number of cells.
Statistical Analysis
Correlation analysis between the area of NETs and other features was conducted with Spearman correlation coefficient (r s). Thrombi areas were excluded because of high density of NETosis. Wilcoxon-Mann-Whitney tests were applied to assess the association between dichotomous traits and the median area of NETs. All statistical tests were two-sided, and P values of < .05 were defined as statistically significant.
Results
Neutrophils Infiltrating the Lungs of Patients With Severe COVID-19
An initial collection of postmortem multiorgan biopsy samples, including lung tissue, from patients with fatal COVID-19 was investigated previously (the exploratory cohort of this study).6 Histologically, a prominent and heterogenous infiltration of macrophages (CD68+) and neutrophils (CD66b+) was found in the lungs, mainly in the interstitium and in areas of diffuse alveolar damage (e-Fig 3A-3D). The number of infiltrating CD4+ T cells (CD3+ CD8–) and CD20+ B lymphocytes varied among the lung tissues. Interestingly, few CD8+ T cells were observed infiltrating all lungs (e-Fig 3D). Overall, an intratissue heterogeneity in the spatial distribution of the leukocyte subsets in the infiltrates was seen.
NETosis in the Lung Tissue from Patients With Fatal COVID-19
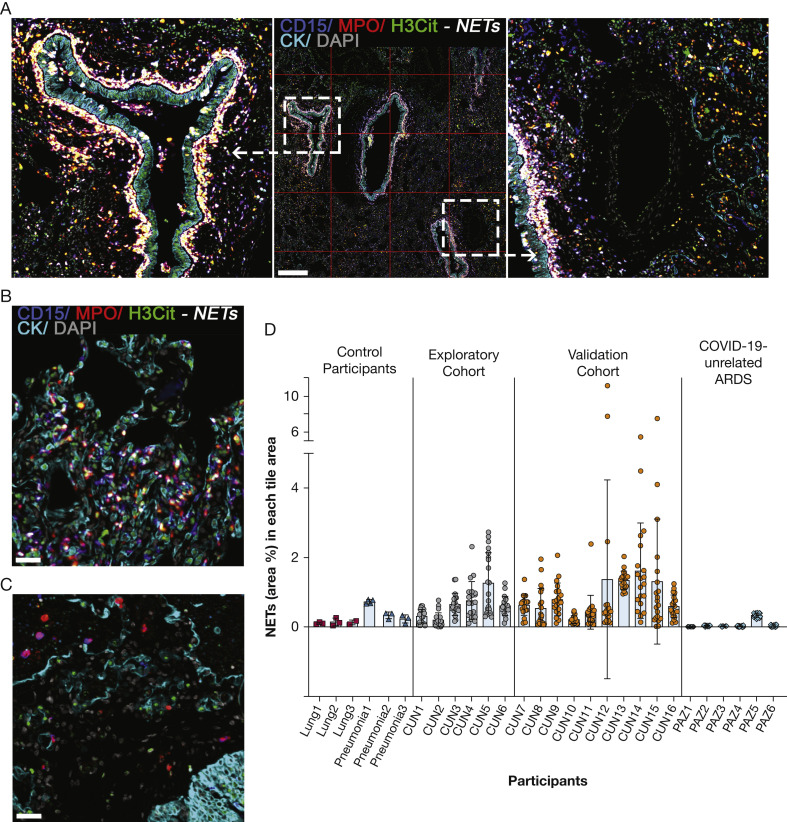
A multiplexed immunofluorescence assay was set up to detect and quantify NETs (Fig 1A-1C; single channels in e-Fig 4; optimization in e-Fig 2). Using serial lung tissue sections and comparing the two multiplexed immunofluorescence assays, a very strong association between NET formation and neutrophil infiltration (CD66b+) was found (r s = 0.941; P < .01) (e-Fig 5). To investigate the global and local spatial distribution of NETs, whole-tissue images were analyzed, and a grid of tiles covering the whole tissue was applied for each image. Each tile had a surface of 1 mm2. NET density varied in the whole-tissue and in each tile area, suggesting both individual and spatial heterogeneity (Fig 1A, 1C; coefficients of variation in e-Table 1). NETs were seen mostly in the lung interstitium, very prominently surrounding the cytokeratin-positive bronchiolar epithelium (Fig 1A, e-Fig 6). Although less frequent, similar events were also seen in bacterial pneumonia (Fig 1B-1D). NETs were detected rarely in healthy lung tissues (Fig 1D, e-Fig 6). Similarly, NETs were hardly detected in the lungs of patients with ARDS unrelated to COVID-19 (Fig 1C-1D, e-Figs 6, 7). NETs seemed to be more common in the lungs because, in liver tissues from the same patients, NETs were found in only one of eight patients whose samples were analyzed thoroughly (e-Fig 8).
Figure 1.
NETs in lung tissues of patients with fatal COVID-19. A, Representative fluorescence image of lung tissue from a patient with fatal COVID-19 showing CD15 (blue channel), MPO (red channel), H3Cit (citrulline R2 + R8 + R17; green channel), cytokeratin (cyan channel), and DAPI (grey channel). NETs are visualized surrounding the bronchiolar epithelium and in the interstitium as white structures by the marker combinations CD15, MPO, and H3Cit. Green channel indicates H3Cit-positive staining of extruded DNA originated from neutrophils undergoing NETosis. A grid of tiles (1 mm2) covering the whole-tissue section was applied in each image to analyze the spatial distribution of NETs. B, Representative fluorescence image of bacterial pneumonia showing scattered NET formation. C, Representative fluorescence image of ARDS unrelated to COVID-19 showing scarce neutrophils (magenta, CD15 [blue channel] plus MPO [red channel]), and no NET formation. D, NET distribution plots showing control tissues (normal lung [n = 6], pneumonia [n = 6]) and lung tissues from patients with fatal COVID-19 (exploratory cohort [n = 6] and validation cohort [n = 10]). Each dot represents the NET area percentage in one tile. Mean values of all tiles are shown. Coefficients of variation are shown in e-Table 2. Fluorescence images for each of the individual markers are shown in e-Fig 4. Scale bars represent 500 μm (A) and 100 μm (B). H3Cit = anti-histone H3; MPO = myeloperoxidase; NET = neutrophil extracellular trap.
NETosis in Lungs With COVID-19 Does Not Correlate With Tissue Viral Loads
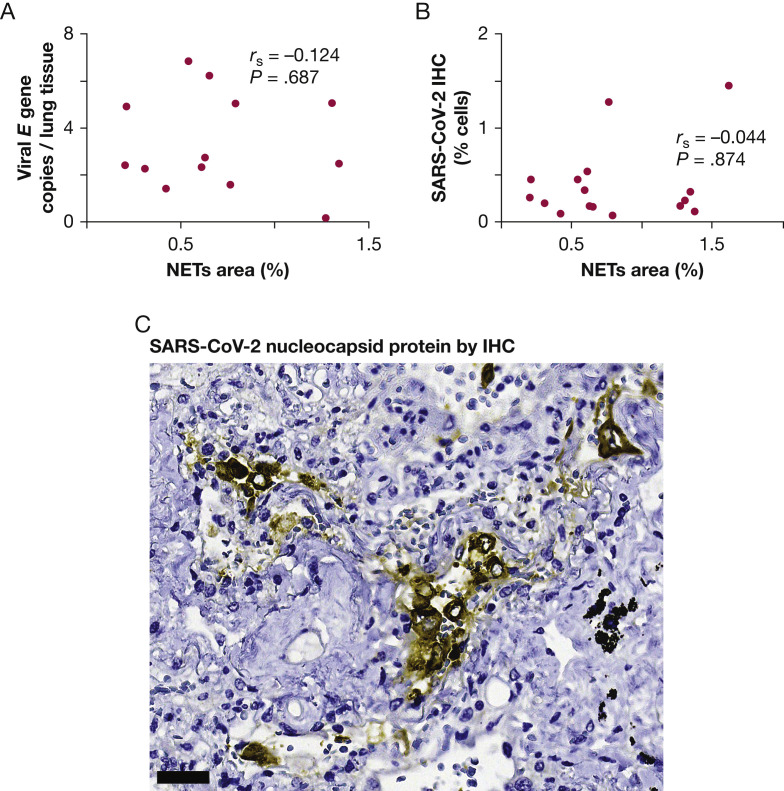
An important question was whether viral presence was determining neutrophil infiltration and NETosis. To explore this possibility, quantitative reverse-transcriptase polymerase chain reactions were used to estimate viral load. No correlation was found between SARS-CoV-2 viral load and NETs in lung tissue (r s = 0.124; P = .687) (Fig 2 A). The number of cells expressing the SARS-CoV-2 N protein was also determined by immunohistochemistry. No correlation was found between SARS-CoV-2 N protein and NETs (r s = –0.044; P = .874) (Fig 2B), suggesting that the viral presence and NET formation in tissue may be independent phenomena. Most of the positive cells morphologically resembled activated macrophages by their size and shape (Fig 2C).
Figure 2.
Lack of association between NETs and the presence of SARS-CoV-2 in lung tissues from patients with fatal COVID-19. A, Scatterplot showing lack of association between the density of NETs (% area) and lung tissue viral loads. B, Scatterplot showing lack of association between NETs and the relative number of cells infected with SARS-CoV-2. C, Photomicrograph showing IHC results for SARS-CoV-2 nucleocapsid protein in large cells that morphologically resemble activated macrophages. Scale bar represents 50 μm. IHC = immunohistochemistry; NET = neutrophil extracellular trap.
Although the presence of neutrophils could be expected to be associated with some patient characteristics, no meaningful correlation with previously diagnosed COPD, endotracheal intubation, length of endotracheal intubation, length of ICU stay, circulating neutrophils, or lymphocytes was found (e-Table 3). Similarly, no associations were observed with circulating D-dimer as indicative of thrombosis and C-reactive protein as indicative of overall systemic inflammatory status. Use of the anti-IL-6R monoclonal antibody tocilizumab also failed to correlate with NET density (e-Table 3). An association between NETs in lung tissue and duration of the clinical course of the disease was observed (r s = 0.49; P < .005) (e-Table 3). Lack of associations may also depend on the various stages of the disease process.
NETosis in Lung Tissue With Fatal COVID-19 Correlates With Focal IL-8 Production and Correlates Inversely With CD8+ T-Cell Infiltration
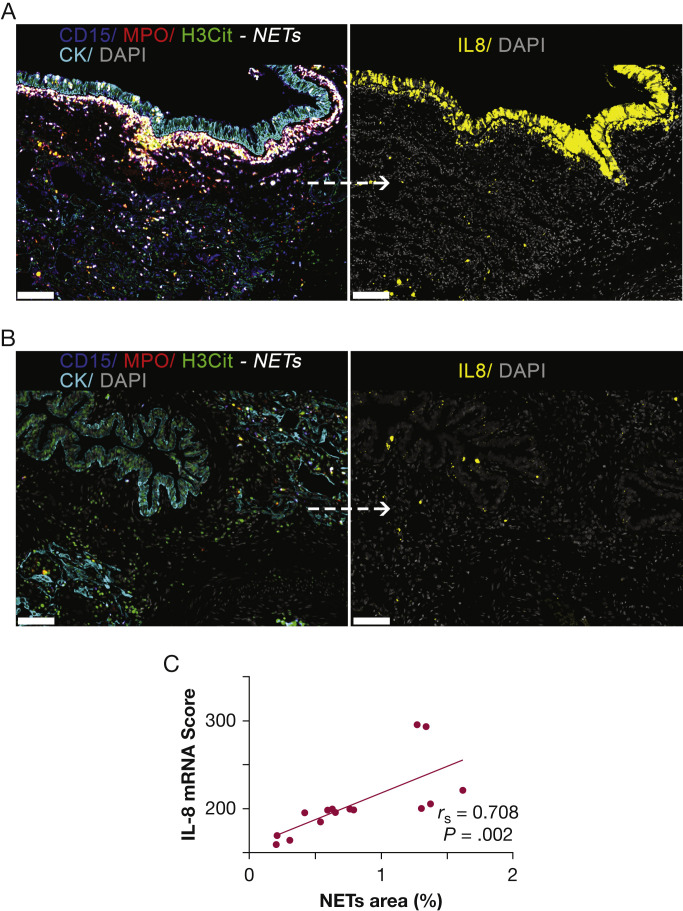
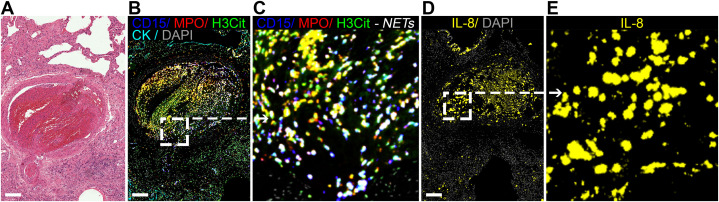
An assay for in situ IL-8 mRNA hybridization was developed by using a human colon cancer (HT-29) cell line as a positive control and a human embryonic kidney 293 (HEK-293) cell line as a negative control (e-Fig 9). The level of IL-8 mRNA was determined in the whole lung tissue sections and then correlated with the local IL-8 protein expression. As expected, an association was found (r s = 0.204; P = .042) (e-Fig 1). Interestingly, IL-8 was highly expressed in some bronchiolar epithelial cells, endothelial cells, and other cell types, likely macrophages and neutrophils (Fig 3A-3B, single channels in e-Fig 10). The level of IL-8 mRNA was associated strongly with the local NET density (r s = 0.708; P = .002) (Fig 3A-3C). Moreover, abundant NETs were found inside thrombi located in lung vessels (Fig 4A-4C, single channels in e-Fig 11), as reported previously.9 , 21 Within the organizing lung thrombi, areas rich in NETs were also found to be enriched in cells transcribing IL-8 (Fig 4B-4E).
Figure 3.
In situ IL-8 mRNA and NETs in lung tissues of patients with fatal COVID-19. A, Representative fluorescence image of lung tissue with a high density of NETs. NETs are visualized especially in the interstitium and surrounding bronchiolar epithelium as white structures by the marker combinations CD15 (blue channel), MPO (red channel), and H3Cit (green channel). Consecutive lung tissue section shows IL-8 mRNA signal (yellow channel) especially seen within and around the bronchiolar epithelium. B, Representative fluorescence image of lung tissue with a very low density of NETs. Consecutive lung tissue section shows a low level of IL-8 mRNA. C, Scatterplot showing the association between NETs and in situ IL-8 mRNA in consecutive lung tissue sections. Fluorescence images for each of the individual markers are shown in e-Fig 10. Scale bars represent 150 μm. H3Cit = anti-histone H3; MPO = myeloperoxidase; mRNA = messenger RNA; NET = neutrophil extracellular trap.
Figure 4.
In situ IL-8 mRNA and NETs in pulmonary thrombi from patients with fatal COVID-19. A, Photomicrograph depicting hematoxylin and eosin stain of lung tissue showing an organizing thrombus in a medium vessel. B, C, Fluorescence images of the pulmonary thrombus showing a high density of NETs. NETs are visualized as white structures by the marker combinations CD15 (blue channel), MPO (red channel), and H3Cit (green channel). D, E, High IL-8 mRNA signal (yellow channel) is found in the organizing thrombus. Fluorescence images for each of the individual markers are shown in e-Fig 11. Scale bars represent 200 μm. H3Cit = anti-histone H3; MPO = myeloperoxidase; mRNA = messenger RNA; NET = neutrophil extracellular trap.
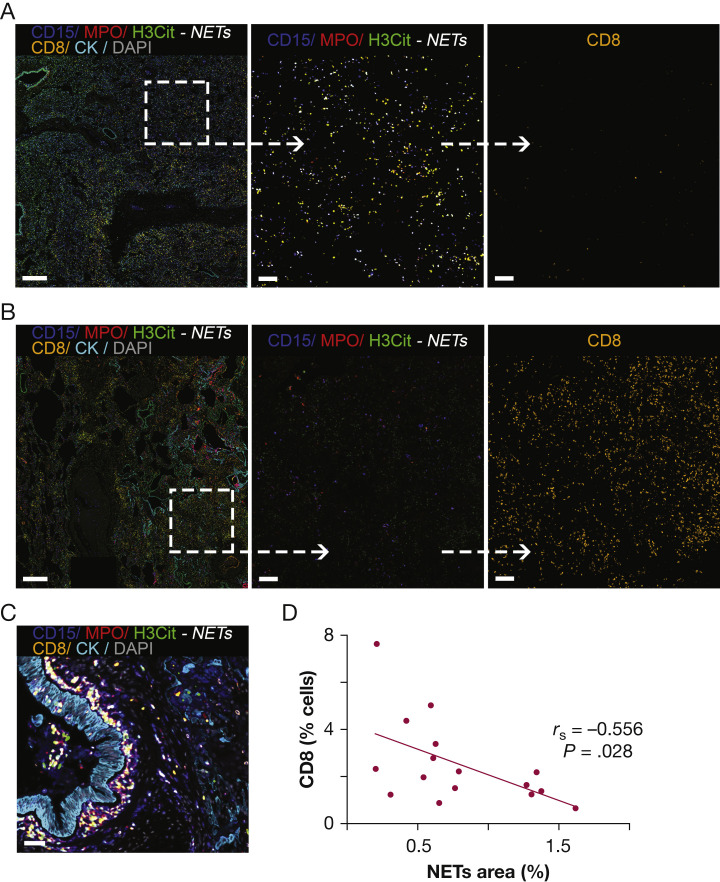
In mouse models of cancer and in human solid tumors, NETs have been shown to correlate negatively with the presence of CD8+ T lymphocytes.22 , 24 This may be relevant in COVID-19 because cytotoxic T lymphocytes are important in the immune responses against RNA viruses. In all lung tissues (exploratory and validation cohorts), a very clear negative correlation between contents of NETs and infiltrating CD8+ T cells was found (Fig 5A-5D, single channels in e-Fig 12). This negative correlation was observed globally, in the whole-tissue analysis, and locally, in the tile analysis (r s = –0.556 for whole-tissue analysis; r s = –0.231 for the tile analysis; P < .01) (Fig 5D). These results suggest that the spatial distribution of areas rich in NETs seemed to be devoid of CD8+ T lymphocytes (Fig 5A-5C). The cytolytic capability of CD8+ T cells was assessed by selective measurement of granzyme B in CD8+ cells. Granzyme B expression was limited to a small subset of CD8+ T cells, representing 1.4% to 9.4% of total CD8+ T cells (e-Fig 13).
Figure 5.
CD8+ T-cell infiltration and NETs in the lung tissue of patients with fatal COVID-19. A, Representative fluorescence image of lung tissue with a high density of NETs. NET-rich areas tend to have low CD8+ T-cell infiltrations. B, Representative fluorescence image of lung tissue with a very low density of NETs. Low neutrophil infiltration (magenta, CD15 [blue channel], MPO [red channel]), and high CD8+ T-cell infiltrations are seen. C, Fluorescence image showing prominent NET formation surrounding peribronchiolar areas with discrete CD8+ T-cell infiltration. D, Scatterplot showing correlation between NETs and CD8+ T cells in whole-tissue sections. Fluorescence images for each of the individual markers are shown in e-Fig 12. Scale bars represent 500 μm (A), 100 μm (B), and 50μm (C). H3Cit = anti-histone H3; MPO = myeloperoxidase; NET = neutrophil extracellular trap.
Discussion
Excessive inflammation is pathogenic in the pneumonitis associated with severe COVID-19. Macrophages and neutrophils are among the most abundantly present leukocytes in the lung inflammatory infiltrates. SARS-CoV-2 infects and replicates in alveolar macrophages that eventually die. Monocyte-derived alveolar macrophages then are recruited, which in turn become infected. This process is likely to sustain local lung inflammation and generates an excessive level of inflammation that ultimately is responsible for tissue damage and organ disfunction.25 This study found a clear neutrophil infiltration in all lung tissues from patients with fatal COVID-19. A spatial heterogeneity of the neutrophil infiltrate distributions was also observed within each lung tissue and may reflect a spectrum from acutely inflamed areas to others with severe lung tissue damage and others with signs of end-stage involvement.
NETs already have been detected in the lungs of patients with fatal COVID-19.18 , 20 , 26 , 27 Herein, we demonstrated, in a relatively large series, that neutrophils and NETs are a constant histopathologic feature in the lungs of patients with fatal COVID-19. Similar to the scattered neutrophil distribution, NETs were also found at variable levels and with a heterogeneous spatial distribution. Their heterogeneous abundance was not linked to tissue viral load. Importantly, the spatial heterogeneity of NETs was associated strongly with local IL-8 levels. In a spatial manner, a negative correlation was found between the abundance of NETs and CD8+ tissue-infiltrating T cells.
NETosis could be the result of several mediators known to be present in COVID-19 pneumonitis such as tumor necrosis factor α, IL-8, IL-1β, C5a, and IL-17A.26 , 28 IL-8 (CXCL8) is a key chemokine for chemotaxis of neutrophils able to elicit NETs in a gradient fashion.29 The clear association between NETs and local IL-8 levels provides circumstantial evidence for involvement of this chemokine in NETosis induction, as reported previously in other disease settings.24 , 30 IL-8 is likely to be both chemoattracting neutrophils and inducing NETosis in COVID-19 pneumonitis. NETosis seems to be more common in the lungs, because NETs were found rarely in the liver tissue from the same patients. As reported previously, liver histologic examination showed discrete features of inflammation.6 This suggests that NET formation seems to be mediated by local inflammation, possibly triggered by IL-8. Similarly, IL-17A has also been shown to induce NETosis in COVID-19.26 Other yet to be discovered factors could also play a role in the observed COVID-associated lung NETosis.
Although SARS-CoV-2 has been shown to induce NETosis directly in an angiotensin converting enzyme 2-dependent fashion,18 this study found no correlation between NETs and tissue viral load. This may reflect the duration of the clinical course and suggests that NETs in the lung of fatal COVID-19 seem to be more related to the strong inflammatory responses than to the direct presence of the virus. Similarly, in a Syrian hamster model, SARS-CoV-2 lung infection also drives NETosis, although it remains to be seen whether through direct or indirect cytokine-mediated mechanisms.31
NETs may play a fundamental role in respiratory failure for their tissue-damaging properties inherent to neutrophil activation. Moreover, the viscosity of extruded DNA could contribute mechanically to reduced lung compliance. Importantly, airways obstructed by NETs were not found, as seen in cystic fibrosis.32 Similar to previous reports,9 , 21 neutrophils and NETs were detected in pulmonary thrombi from patients with fatal COVID-19. Importantly, IL-8 secreted within the organizing thrombi may chemoattract neutrophils potently and may induce NET formation, potentially linking inflammation and thrombosis.27
If considered pathogenic, it would be logical to interfere with the NETosis pathway by pharmacologic routes. Drugs inhibiting the peptidyl arginine deiminase 4 enzyme inhibit NETosis.33 Moreover, anti-IL-8 monoclonal antibodies are being tested in clinical trials,34 , 35 and CXCR1 and CXCR2 pharmacologic inhibitors are available.36 In addition, anti-IL-17A antibodies are also used in patients with psoriasis, and IL-17A has been shown to elicit NETosis in experimental pancreatic cancer.37 In the case of IL-8, this issue is especially important because alternative chemokines are known to elicit NETosis acting on the same receptors such as CXCL1 and CXCL2. Thus, tampering with receptor function could be the best option. Direct interventions to degrade extracellular DNA with DNase I are also possible, and clinical trials with inhalation delivery of clinical-grade DNAse I (Pulmozyme, Genentech) are in progress.38 , 39 However, penetration of the enzyme into the interstitium where we observed the NETs, particularly around the airways, may be an issue.
Interpretation
Our results showed that NETosis is a prominent and constant histopathologic feature of severe COVID-19 in the lungs, and that this seems to correlate with IL-8 expression and with weaker CD8 T-cell presence in the tissue. The mechanisms whereby NETs could interfere with CD8 T lymphocytes remain to be elucidated. Importantly, CD8+ T-cell responses are likely to contribute to virus clearance.40 Causative associations remain to be addressed, but NETs and NET formation offer therapeutically actionable targets already under investigation.
Acknowledgments
Author contributions: I. M., M. V.-E., A. T., L.-P. H., P. K., F. I., M. F. L, and C. E. d. A. conceived the study. M. V.-E., B. R.-Z., A. A., L. G.-T., L. A.-G., M. A.-M, M. F.-A., G. R., F. C.-T., J. A. Q., J. L. D. P, A. C., J. I. E., A. L.-J., D. H., M. D. L., L.-P. H., P. K., F. I., M. F. L., and C. E. d. A. designed the experiments. M. V.-E., D. J. S., B. R.-Z, A. A., L. G.-T., L. A.-G., C. S., D. G.-R., E. T., M. R.-M., G. R., and A. C. generated and analyzed the data and contributed to data interpretation. I. M., M. V.-E., J. I. E., M. D. L., L.-P. H., P. K., F. I., M. F. L., and C. E. d. A. wrote the manuscript with input from all authors.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following: I. M. reports receiving commercial research grants from BMS, Bioncotech, Alligator, Pfizer, Leadartis, and Roche; has received speakers bureau honoraria from MSD; and is a consultant or advisory board member for BMS, Roche, Genmab, F-Star, Bioncotech, Bayer, Numab, Pieris, Alligator, and Merck Serono. C. E. d. A. reports research grants from AstraZeneca. F. I. is a Wellcome Trust CRCD Fellow 211122/Z/18/Z. L.-P. H. is supported by the Oxford NIHR Biomedical Research Centre. P. K. is supported by NIH U19 AI082630. None declared (M. V.-E., B. R.-Z., D. J.-S., A. T., A. A., L. G.-T., L. A.-G., C. S., D. G.-R., E. T., M. A.-M, M. F.-A., M. R.-M., G. R., F. C.-T., J. A. Q., J. L. D. P, A. C., A. L.-J., D. H., J. I. E., M. D. L., M. F. L.).
Funding/support: This study was funded by the Banco Bilbao Vizcaya (BBVA) Foundation, “Ayudas a Equipos de Investigación Científica SARS-CoV-2 y COVID-19.”
Role of sponsors: The sponsor had no role in the design of the study, the collection and analysis of the data, or the preparation of the manuscript.
Other contributions: The authors thank the patient families and the nursing staff of the Clínica Universidad de Navarra, and Drs Tiong Kit Tan, PhD, and Alain Townsend, PhD, of University of Oxford and Dr Kuan-Ying A. Huang, PhD, of Chang Gung University for the antibody production for SARS-CoV-2 detection. F. I. and A. C. acknowledge the philanthropic support of the donors to the University of Oxford’s COVID-19 Research Response Fund.
Additional information: The e-Figures and e-Tables are available online under “Supplementary Data.”
Footnotes
Drs Melero and Villalba-Esparza contributed equally to this manuscript.
Supplementary Data
References
- 1.Islam N., Shkolnikov V.M., Acosta R.J., et al. Excess deaths associated with covid-19 pandemic in 2020: age and sex disaggregated time series analysis in 29 high income countries. BMJ. 2021;373:n1137. doi: 10.1136/bmj.n1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baud D., Qi X., Nielsen-Saines K., Musso D., et al. Real estimates of mortality following COVID-19 infection. Lancet Infect Dis. 2020;20:773. doi: 10.1016/S1473-3099(20)30195-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin D.-Y., Gu Y., Wheeler B., et al. Effectiveness of Covid-19 vaccines over a 9-month period in North Carolina. N Engl J Med. 2022;386:933–941. doi: 10.1056/NEJMoa2117128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merad M., Blish C.A., Sallusto F., Iwasaki A. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 5.Nouailles G., Wyler E., Pennitz P., et al. Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19. Nat Commun. 2021;12:4869. doi: 10.1038/s41467-021-25030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Recalde-Zamacona B., García-Tobar L., Argueta A., et al. Histopathological findings in fatal COVID-19 severe acute respiratory syndrome: preliminary experience from a series of 10 Spanish patients. Thorax. 2020;75:1116–1118. doi: 10.1136/thoraxjnl-2020-215577. [DOI] [PubMed] [Google Scholar]
- 7.Laing A.G., Lorenc A., Del Molino Del Barrio I., et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 8.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217 doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackermann M., Verleden S.E., Kuehnel M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulte-Schrepping J., Reusch N., Paclik D., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szabo P.A., Dogra P., Gray J.I., et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardhana S.A., Wolchok J.D. The many faces of the anti-COVID immune response. J Exp Med. 2020;217 doi: 10.1084/jem.20200678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teijeira A., Garasa S., Ochoa M.C., et al. Interleukin-8, neutrophils, and NETs in a collusion against cancer immunity and immunotherapy. Clin Cancer Res. 2020;27:2383–2393. doi: 10.1158/1078-0432.CCR-20-1319. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann V., Reichard U., Goosmann C., et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 15.Branzk N., Lubojemska A., Hardison S.E., et al. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat Immunol. 2014;15:1017–1025. doi: 10.1038/ni.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keir H.R., Chalmers J.D. Neutrophil extracellular traps in chronic lung disease: implications for pathogenesis and therapy. Eur Respir Rev. 2022;31 doi: 10.1183/16000617.0241-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keir H.R., Shoemark A., Dicker A.J., et al. Neutrophil extracellular traps, disease severity, and antibiotic response in bronchiectasis: an international, observational, multicohort study. Lancet Respir Med. 2021;9:873–884. doi: 10.1016/S2213-2600(20)30504-X. [DOI] [PubMed] [Google Scholar]
- 18.Veras F.P., Pontelli M.C., Silva C.M., et al. SARS-CoV-2-triggered neutrophil extracellular traps mediate COVID-19 pathology. J Exp Med. 2020;217 doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouwendijk W.J.D., Raadsen M.P., van Kampen J.J.A., et al. High levels of neutrophil extracellular traps persist in the lower respiratory tract of critically ill patients with coronavirus disease 2019. J Infect Dis. 2021;223:1512–1521. doi: 10.1093/infdis/jiab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radermecker C., Detrembleur N., Guiot J., et al. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J Exp Med. 2020;217 doi: 10.1084/jem.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leppkes M., Knopf J., Naschberger E., et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Andrea C.E., Ochoa M.C., Villalba-Esparza M., et al. Heterogenous presence of neutrophil extracellular traps in human solid tumours is partially dependent on IL-8. J Pathol. 2021;255:190–201. doi: 10.1002/path.5753. [DOI] [PubMed] [Google Scholar]
- 23.Abengozar-Muela M., Esparza M.V., Garcia-Ros D., et al. Diverse immune environments in human lung tuberculosis granulomas assessed by quantitative multiplexed immunofluorescence. Mod Pathol. 2020;33:2507–2519. doi: 10.1038/s41379-020-0600-6. [DOI] [PubMed] [Google Scholar]
- 24.Teijeira Á., Garasa S., Gato M., et al. CXCR1 and CXCR2 chemokine receptor agonists produced by tumors induce neutrophil extracellular traps that interfere with immune cytotoxicity. Immunity. 2020;52:856–871. doi: 10.1016/j.immuni.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Grant R.A., Morales-Nebreda L., Markov N.S., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azevedo M.L.V., Zanchettin A.C., Vaz de Paula C.B., et al. Lung neutrophilic recruitment and IL-8/IL-17A tissue expression in COVID-19. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.656350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Middleton E.A., He X.-Y., Denorme F., et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vabret N., Britton G.J., Gruber C., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teijeira A., Garasa S., Ochoa M.D.C., et al. Differential Interleukin-8 thresholds for chemotaxis and netosis in human neutrophils. Eur J Immunol. 2021;51:2274–2280. doi: 10.1002/eji.202049029. [DOI] [PubMed] [Google Scholar]
- 30.Schauer C., Janko C., Munoz L.E., et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med. 2014;20:511–517. doi: 10.1038/nm.3547. [DOI] [PubMed] [Google Scholar]
- 31.Becker K., Beythien G., de Buhr N., et al. Vasculitis and neutrophil extracellular traps in lungs of golden Syrian hamsters with SARS-CoV-2. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.640842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skopelja S., Hamilton B.J., Jones J.D., et al. The role for neutrophil extracellular traps in cystic fibrosis autoimmunity. JCI Insight. 2016;1 doi: 10.1172/jci.insight.88912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis H.D., Nacht M. iPAD or PADi-‘tablets’ with therapeutic disease potential? Curr Opin Chem Biol. 2016;33:169–178. doi: 10.1016/j.cbpa.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 34.National Institutes of Health Clinical Center. A study of BMS-986253 in combination with nivolumab or nivolumab plus ipilimumab in advanced cancers. NCT03400332. ClinicalTrials.gov. National Institutes of Health; 2018. Updated May 18, 2022. https://clinicaltrials.gov/ct2/show/NCT03400332
- 35.National Institutes of Health Clinical Center. Combination Study of AZD5069 and Enzalutamide (ACE). NCT03177187. ClinicalTrials.gov. National Institutes of Health; 2017. Updated March 21, 2022. https://clinicaltrials.gov/ct2/show/NCT03177187.
- 36.Ruffini P.A. The CXCL8-CXCR1/2 axis as a therapeutic target in breast cancer stem-like cells. Front Oncol. 2019;9:40. doi: 10.3389/fonc.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Chandra V., Riquelme Sanchez E., et al. Interleukin-17-induced neutrophil extracellular traps mediate resistance to checkpoint blockade in pancreatic cancer. J Exp Med. 2020;217 doi: 10.1084/jem.20190354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Institutes of Health Clinical Center. Pulmozyme to Improve COVID-19 ARDS Outcome. NCT04402944. ClinicalTrials.gov. National Institutes of Health; 2020. Updated December 17, 2021. https://clinicaltrials.gov/ct2/show/NCT04402944
- 39.National Institutes of Health Clinical Center. Nebulised dornase alfa for treatment of COVID-19 (COVASE). NCT04359654. ClinicalTrials.gov. National Institutes of Health; 2020. Updated November 9, 2021. https://clinicaltrials.gov/ct2/show/NCT04359654.
- 40.Kroemer M., Spehner L., Vettoretti L., et al. COVID-19 patients display distinct SARS-CoV-2 specific T-cell responses according to disease severity. J Infect. 2021;82:282–327. doi: 10.1016/j.jinf.2020.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.