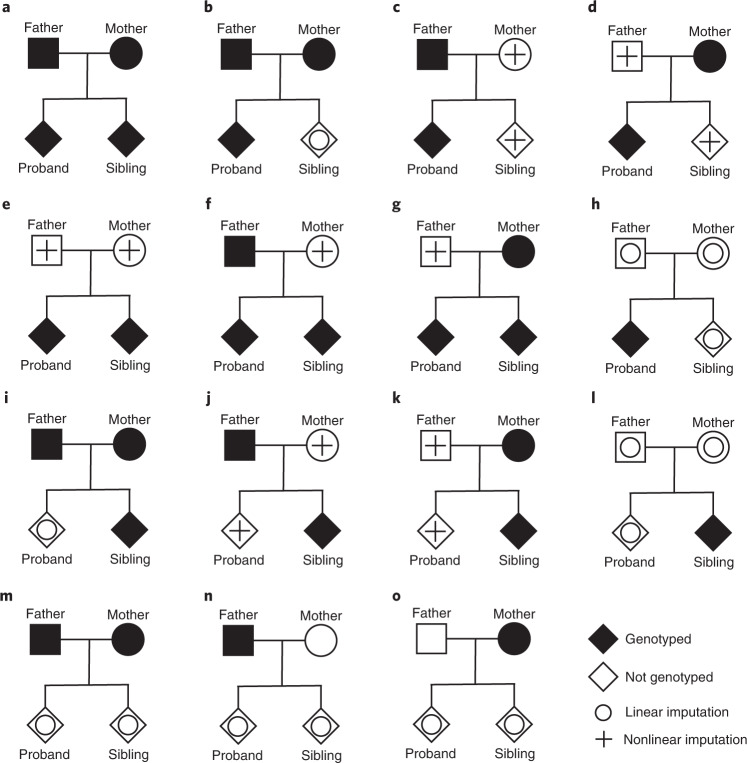
Fig. 1. Mendelian imputation for 15 different missing genotype cases.
Displayed are cases where one offspring is phenotyped (the proband), but our framework can handle cases where both offspring are phenotyped. We distinguish between imputations that are linear functions of observed genotypes and imputations that are nonlinear functions of observed genotypes, and thereby add information for estimating the parameters of models (1) and (2). There are seven cases (c–g, j and k) where nonlinear imputations are possible, but we note that in some of these cases (c–e, j and k), the resulting variance-covariance matrix of the observed and imputed genotypes is not of full rank (Extended Data Fig. 1), implying that the full parameter vector of model (2) cannot be identified based on data of that type alone. For example, in case e, the imputed paternal and maternal genotypes are the same, but the imputed sum of paternal and maternal can be used to estimate the parameters of model (3). We detail how to combine information from different data types below. Although we show the case of two offspring here to simplify exposition, our imputation method and software (snipar) can handle any number of genotyped offspring (Methods and Supplementary Note Sections 3 and 5).