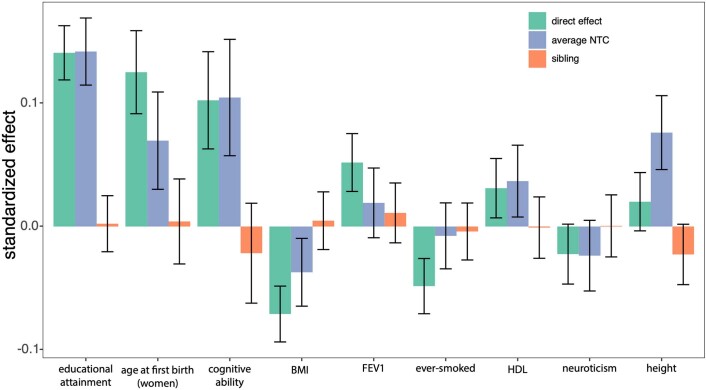
Extended Data Fig. 5. Indirect sibling effects of the EA polygenic index.
The standardized effect estimate (SD change in trait per SD change in PGI) is given along with the 95% confidence interval. Estimates were derived from a sample of 35,556 individuals from the ‘White British’ subsample of the UKB with at least one genotyped sibling and imputed and/or observed parental genotypes. Effects were estimated by joint regression of individuals’ traits onto their own PGI, their siblings’ PGI, and their mother’s and father’s (imputed/observed) PGI (Methods). We give the average of the maternal and paternal NTCs, adjusted for bias due to imputation in the presence of assortative mating (Methods), as the ‘average NTC’ here. Traits were adjusted for 40 genetic principal components prior to analysis. Trait abbreviations: HDL, high-density lipoprotein cholesterol; BMI, body mass index; FEV1, forced expiratory volume in one second.