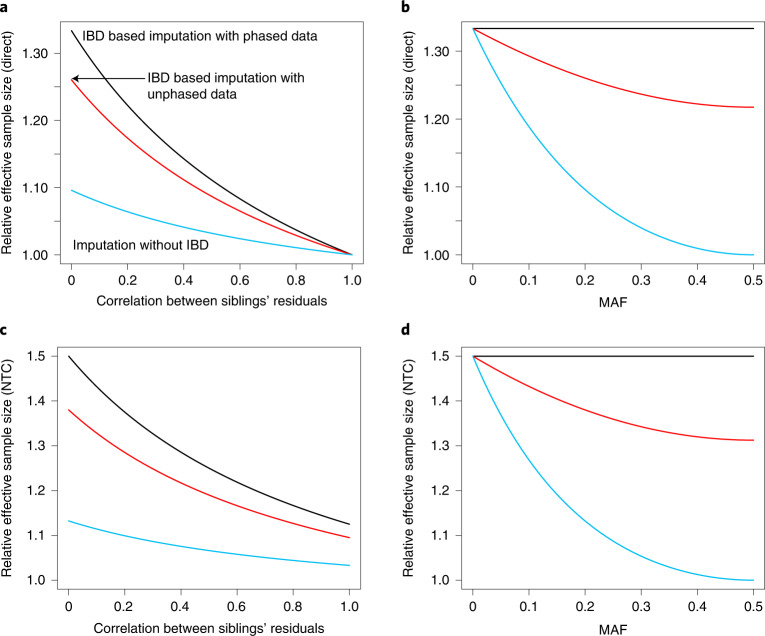
Fig. 3. Relative efficiency for estimation of direct effects and NTCs from sibling pairs using different imputation methods.
We compare the theoretical effective sample size for estimation of direct genetic effects and average NTCs from three imputation methods: one that does not use IBD segments (blue)39, one that uses IBD and unphased data (red) and one that uses IBD and phased data (black). Effective sample size is measured relative to that from using sibling genotypes alone without any imputation and assuming that we have a sample of independent families with two genotyped and phenotyped siblings in each family (Supplementary Note Section 4). a, Effective sample size for estimation of the direct genetic effect when MAF is 20% as a function of correlation between siblings’ residuals. b, Effective sample size for estimation of direct genetic effects as a function of MAF when the correlation between siblings’ residuals is zero. (Results follow a similar pattern for other sibling correlations.) For imputation from unphased data, when both siblings are heterozygous and share one allele IBD, which allele is shared IBD cannot be determined (Fig. 2b), so the imputation averages over the two possibilities. When phased data are used, the observed parental alleles can be determined, so the relative efficiency does not depend upon MAF. c, The same as for a, but for average NTC. d, The same as for b, but for average NTC.