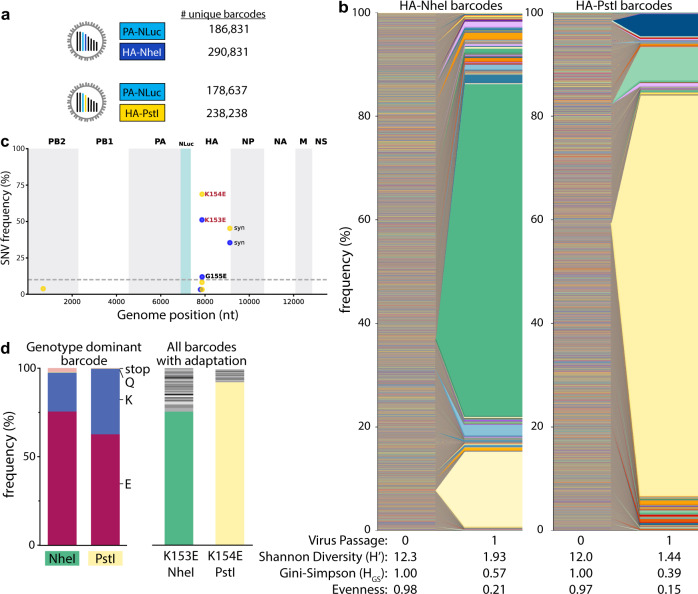
Fig. 2. Single viruses seed selective sweeps in HA.
a Creation of two dual-barcode libraries with distinct registration marks and uniquely addressable members. b Frequency of each lineage as a fraction of total population size. Colors indicate unique barcode identities. c Whole-genome sequencing identifies adaptive variants in HA. Individual single nucleotide variant (SNV) frequencies are indicated at each nucleotide position in a concatenated IAV genome for each library. d Long-read sequencing reveals selective sweeps by linking adaptive mutants in HA to single dominant barcodes. The frequencies of mutations coding for the indicated change that is linked to the dominant barcode are shown for both libraries (left). The actual frequency of K > Q and K > stop mutants versus the contribution from long-read sequencing errors to their appearance is unknown. The frequencies of all barcodes linked to the adaptive glutamic acid variant are indicated (right), with the dominant barcode in each library colored as in b. Source data are provided as a Source Data file.