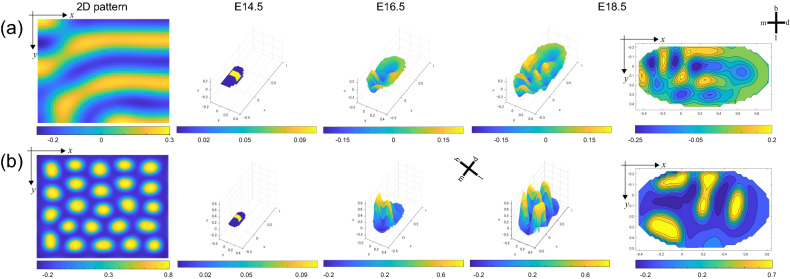
Figure 4.
Simulation of molar morphogenesis. The parameters for UM that make stripe patterns are the following: (a) fu = 0.4, fv = − 1, q = 0, c = 1, gu = 0.4, gv = − 0.44, du = 0.006, dv = 0.06. The parameters for LM that make spot patterns are the following: (b) fu = 0.2, fv = − 0.6, q = 0.45, c = 0.6, gu = 0.2, gv = − 0.32, du = 0.006, dv = 0.06. The 2D images of stripe and spot patterns are formed after letting the system settle from an initially random configuration under the above parameter sets. Simulated concentration of activator (u) is represented in 3D at E 14.5, E 16.5, and E. 18.5. The final plain colormaps are also shown at E 18.5. The unit of x and y axis is in mm. m mesial, d distal, b buccal, l lingual.