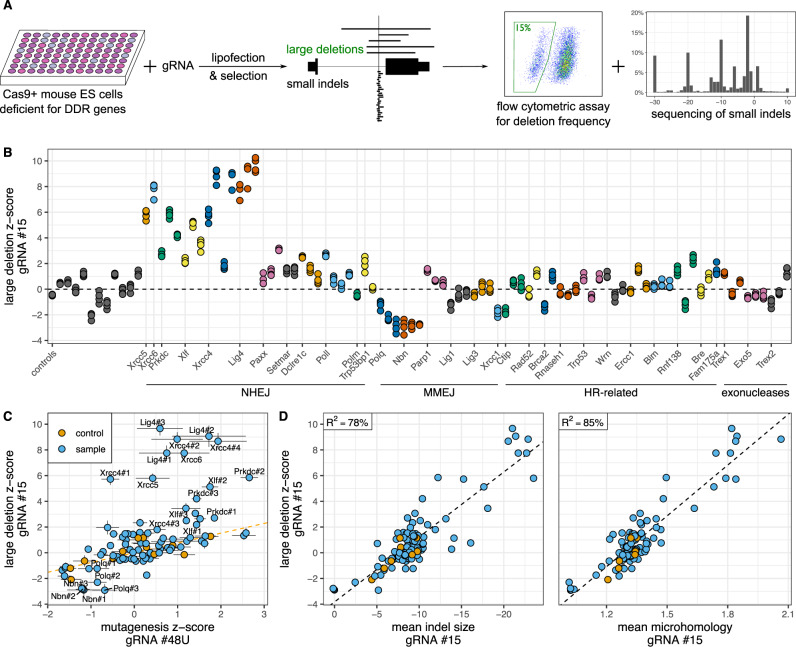
Fig. 1. End joining pathways divergently control the frequency of large deletions caused by Cas9 in mouse ES cells.
A Experimental design. Library of Cas9-positive NGS-validated clones deficient for DNA damage repair genes was transfected with individual gRNA-expressing constructs and selected for stable integration. Expression of target genes was measured by flow cytometry, revealing frequency of large deletions (using intronic gRNA #15) or overall mutagenesis (using gRNAs #48U, #48 and #148). Frequency of small indels was established by targeted sequencing of short-range PCR products. B Frequency of large deletions caused by Cas9 with intronic gRNA in DNA damage deficient clones, measured by flow cytometry, expressed as a regressed z-score (see Methods). Only the initial clone in a series of clones deficient for the same gene is labeled on the x axis. N = 4 independent cell cultures. C Comparison of large deletion and mutagenesis indices (see Methods). Dashed line indicates best linear fit to control clones (in orange). Error bars are 2xSEM (N = 4). D Correlation between large deletion z-score (measured by flow cytometry) and the size or microhomology extent of small indels (measured by targeted sequencing). Each dot represents an average readout of an individual clone (N = 1–2 biologically independent cell cultures). Negative indel sizes indicate dominance of deletions.