Figure 3.
Evaluation of the cardiomyocyte differentiation potential of the genome-edited iPSCs
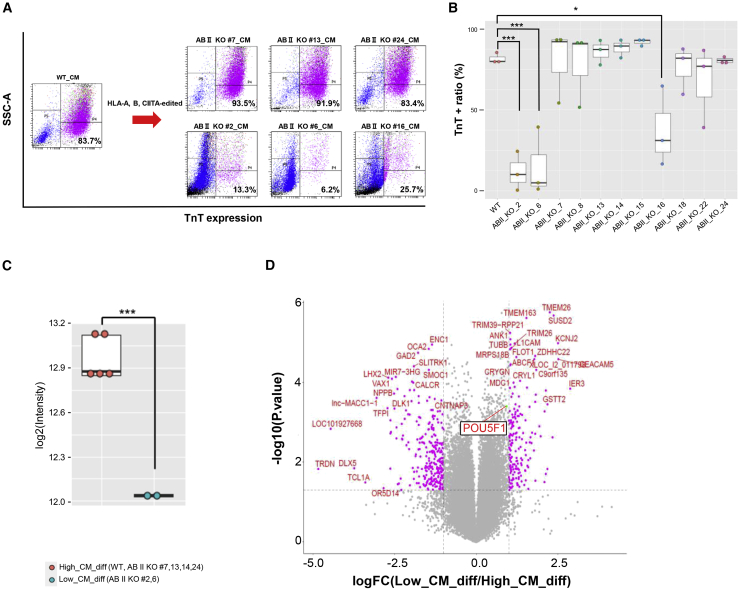
(A) Scatterplots of the FCM analysis examining the proportion of troponin T (TnT)+ cells at 15 days after the induction of cardiomyocyte differentiation. The results of the original (wild-type) and typical results of each three clones showing high or low cardiomyocyte differentiation potential. The results of other clones are shown in Figure S3A. The horizontal axis indicates the fluorescence intensities of fluorophores conjugated with the TnT antibody. The vertical axis indicates side scatter. (B) A boxplot showing the proportion of TnT+ cells in cardiomyocyte differentiation assays that were performed on three biological replicates. Asterisks indicate statistical significance: ∗∗∗p < 0.001; ∗p < 0.05. (C) A boxplot of microarray data showing the differential expression of POU5F1 in iPSC clones showing high (orange) or low (cyan) cardiomyocyte differentiation potential. Each dot shows a clone. Asterisks indicate statistical significance: ∗∗∗p < 0.001. (D) A volcano plot of the microarray analysis showing the differentially expressed genes between iPSC clones with high or low cardiomyocyte differentiation potential.