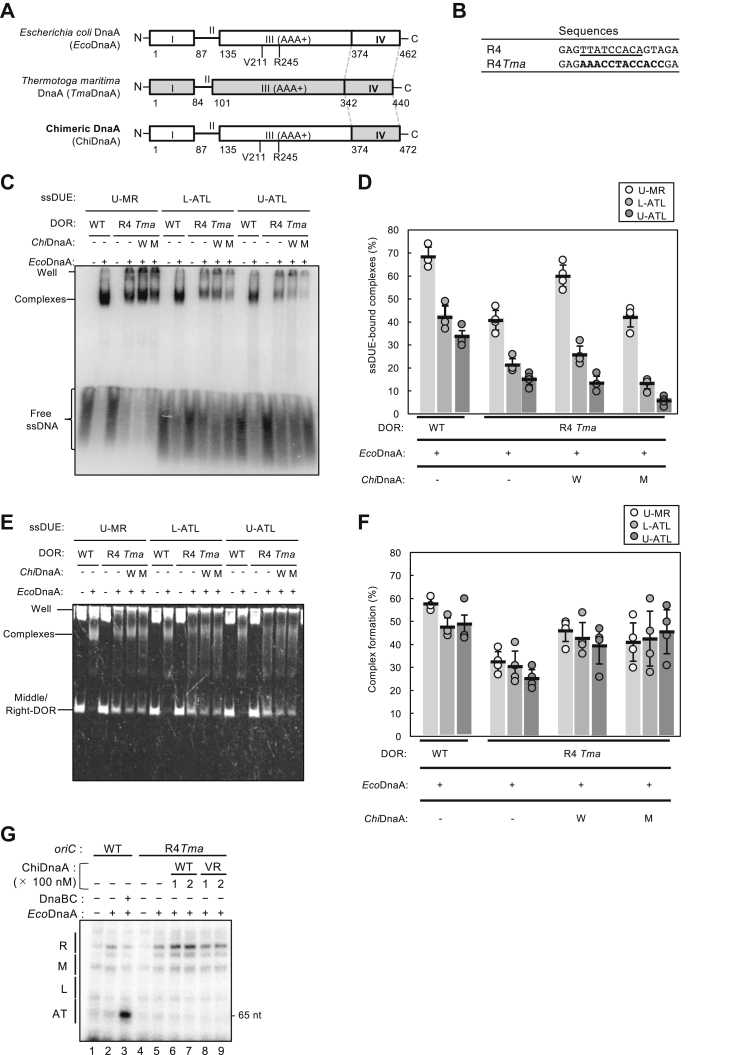
Figure 4.
Role for DnaA box R4 in ssDUE bindingby Middle/Right-DOR–DnaA complex. A, basic structure of ChiDnaA. Schematic structures of Escherichia coli DnaA (EcoDnaA), Thermotoga maritima DnaA (TmaDnaA), and ChiDnaA are shown. Domains of EcoDnaA and TmaDnaA are indicated by open and gray boxes, respectively. B, substitution of DnaA box R4. Sequences of original oriC R4 box (underline) and substituted TmaDnaA box (bold letters) are shown. In R4Tma: oriC, the TmaDnaA box replaces the R4 box. C–F, EMSA using the Middle/Right-DOR with or without substitution of the TmaDnaA box for the R4 box. Middle/Right-DOR WT or R4Tma (35 nM) was incubated for 5 min on ice in the presence (120 nM) (+) or absence (−) of ATP-EcoDnaA, ATP-ChiDnaA WT (W), or ATP–ChiDnaA V211A/R245A (M), followed by further incubation for 10 min at 30 °C with 32P-labeled ssDNA U-MR, U-ATL, or L-ATL (16 nM) in buffer including 100 mM potassium glutamate (see Experimental procedures). Resultant complexes were analyzed by radioactive imaging or GelStar staining, respectively (C and E). The amounts of ssDNA bound to the DnaA–DOR complexes were quantified as “ssDNA-bound complex (%)”(D). Band intensities of Middle/Right-DOR–DnaA complexes were quantified as “Complex formation (%)” (F). Three to four independent experiments were carried out, and each data and mean values with SDs (n = 3–4) are shown in the graphs. When EcoDnaA and the DORs bearing the R4Tma substitution were coincubated, abnormal complexes such as aggregates may have formed, including λ DNA, resulting in material remaining in the gel wells. G, KMnO4 modification experiments. Plasmid bearing oriC (5 nM) with the WT R4 box (WT) or the substituted TmaDnaA box sequence (R4Tma) were incubated for 10 min at 37 °C with IHF (100 nM) in the presence (+) or absence (−) of EcoDnaA (100 nM), ChiDnaA WT or V211A/R245A (VR) (100 or 200 nM), DnaB K236A (300 nM), and DnaC (300 nM), followed by further incubation with 10 mM KMnO4. Modified DNAs were analyzed by primer extension and 7.5% denaturing mini-gel PAGE. Sequencing reactions were performed using the same primer, and determined positions of sequence motifs (L, M, R, and AT) are shown using the oriC WT KMnO4 modification data (Fig. S3). Similar results were shown in repeated experiments. DUE, duplex unwinding element; ChiDnaA, chimeric DnaA; EcoDnaA, Escherichia coli DNA; TmaDnaA, Thermotoga maritima DnaA; ss, single stranded.