Abstract
Neuraminidase (NA) is a second major surface protein of the influenza virus and has recently been suggested as a supplemental antigen to the major immunodominant hemagglutinin (HA) antigen in the influenza vaccine. NA is less affected by antigenic drift compared to the HA, induces strong anti-neuraminidase immune responses, and provides broader protection against many influenza strains. However, the NA amount in currently licensed influenza virus vaccines is much lower than that of HA, and not standardized. A platform to produce NA antigen, in the form of virus-like particles (VLPs), was thus developed, to facilitate supplementation of NA antigen in the influenza vaccine formula. Stably transformed Sf9 insect cells had been engineered to express the influenza A virus (H5N1) NA gene under a baculovirus OpMNPV IE2 promoter. Recombinant NA protein was synthesized and assembled into VLPs, in the intact cellular environment provided by insect cells. Approximately 150 µg/ml of NA-VLPs was obtained in the culture medium. Purification of the NA-VLPs was achieved by a sucrose density gradient ultracentrifugation. The purified NA-VLPs effectively induced anti-NA antibodies with neuraminidase inhibition activities in mice. This work demonstrates a simple process to produce an immunocompetent NA-VLPs antigen, exclusively made of only neuraminidase, by insect cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12033-022-00519-8.
Keywords: Influenza A virus, Influenza vaccine, Neuraminidase, Sf9, Stably transformed insect cell, NA-VLPs
Introduction
Influenza A virus causes acute respiratory illnesses and is still a major threat to global public health. Hemagglutinin (HA) and neuraminidase (NA) are two major viral surface glycoproteins. It was estimated that the influenza virus contains approximately 25–30% HA and only 1.5–8% NA out of total virus proteins [1–4]. HA exists as a trimer that binds to sialic acid residues links to glycoproteins and glycolipids of the receptor on the host cell surface and triggers virus internalization by endocytosis [5]. Currently licensed influenza vaccines are therefore use HA as a major antigen to induce anti-HA antibodies for influenza virus protection. However, HA is constantly evolving, causing antigenic drift and occasional antigenic shift in influenza type A viruses. Annual reformulation and re-administration of influenza vaccines are thus recommended [6, 7]. The second surface protein NA, on the other hand, has 90% homology between the strains within the same subtype, 50% homology between subtypes, and slower antigenic evolution compared to the HA [8]. The NA is a tetrameric type II transmembrane protein with an enzymatic function that destroys the receptor by cleaving terminal sialic acid from its glycans on the host cell surface receptors [9, 10]. Type A influenza NA is a three-domain protein [11]. The largest domain is the enzymatic head, which is brought to the viral membrane by a filamentous stalk domain connected to an N-terminal transmembrane domain (TMD) [12]. TMD forms a homo-tetramer and efficiently traffics to the plasma membrane where it along with the HA, matrix 2 protein, eight distinct influenza virus ribonucleoproteins, and other viral components are packaged into budding virions [13]. Despite their antagonistic activities, competitive cooperation between NA and HA during virus entry was suggested [14]. NA may facilitate the virus to move across airway mucus to access the right HA receptors [15, 16]. Another role of NA that has been well demonstrated is when the virus buds from infected cells. It enables the efficient release of the nascent viral particles from the host cell by cleaving off terminal sialic acids. Antibody to NA can block the release of progeny viruses from the host cell [17]. Individuals with high anti-NA antibody titers had reduced virus shedding and illness after infection by both matched and cross-protection against influenza virus strains [18–21]. Despite the benefits of anti-NA antibody, commercial inactivated and split influenza vaccines only induce low and different levels of NA-specific immune response [22, 23]. This is due to low content of the NA in the virus particle compared to HA as previously described and different manufacturing processes, respectively, leading to limited and variable NA immunogenicity [1, 24]. Thus, vaccines that induce robust and protective anti-NA immunity should be developed.
One promising immunogenic form for presenting the NA antigen for this purpose is a multimeric viral structural protein assembled into virus-like particle (VLP). VLPs take advantage of similarity in morphological to the real virus, therefore, providing strong immunogenicity. Since there is no virus genetic material in the VLP, it constitutes a safe alternative to inactivated and live-attenuated influenzas virus antigens. Influenza VLPs are traditionally composed of hemagglutinin (HA) protein displayed on the surface of the M1 protein (the scaffold). However, Lai et al. [25] demonstrated that the NA of both human subtypes of neuraminidase, N1 and N2, could form virus-like particles (VLPs) when expressed individually from plasmid DNA without the presence of hemagglutinin (HA) or M1 protein [25]. Hence, the NA-VLPs could be entirely made from the NA, which is a high potential form for an NA-based influenza vaccine. An efficient, safe, simple, and scalable manufacturing process for NA-VLP production in sufficient amounts should therefore be developed. Insect cells are a promising choice for this purpose. Insect cells not only are well-accepted safety profiles due to their minimal potential for growth of human pathogens, but also have abilities to perform eukaryote post-translational modifications. More importantly, they can be grown as suspension cells in simple culture processes and hence running batch and fed-batch bioreactors on large scale are straightforward. The NA gene could be delivered and be expressed by the insect cells using expression vectors, e.g., plasmid and virus vectors such as baculovirus. Although baculovirus is the most widely used expression vector for insect cells, baculovirus infection causes some adverse effects on insect cell secretory pathway [26] and hence lower yields and qualities of the secreted protein. In addition, extra steps for the removal of baculovirus and cellular proteins from infected cell lysis must be included in the purification process. Thus, a plasmid-driven gene expression was chosen for the development of stably transformed insect cell lines, which had the NA gene integrated into the insect chromosome, and expressed under the control of a baculovirus (OpMNPV) IE-2 promoter [27]. Since there is no virus infection involved, the plasmid transformed insect cells have full capacity for post-translational modifications, continuity of protein production and high yields of secreted recombinant proteins. It was anticipated that the recombinant NA protein synthesized by these cells would be assembled into VLPs and secreted out of the cells. Upstream and downstream processes developed for this NA antigen are discussed.
Materials and Methods
Insect Cell Culture
Spodoptera frugiperda (Sf9 cell line, ATCC CRL-1711) were maintained in a modified Grace’s medium (Gibco, Thermo Fisher Scientific, Inc., USA) supplemented with 10% fetal bovine serum (FBS). Sf9 cells were added to a flask with 20 ml of fresh complete medium for a final density of 3 × 105 viable cells/ml and placed in an incubator shaker set at 27 °C with the shaking speed at 120 rpm for 3 days. Sub-culture was done when the cell reached the mid-log phase of growth. In this study, the Sf9 cells were also gradually adapted to grow in a serum-free medium, Sf900-II (Thermo Fisher Scientific, Inc., USA), using similar culture conditions.
Plasmid Construction
Influenza A virus [A/Thailand/1(KAN1)/2004/H5N1] neuraminidase gene (NA, GenBank no. AY555151.3) was kindly provided by Professor Pilaipan Puthavathana, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand. The full-length NA DNA fragment was digested by KpnI and EcoRI restriction enzymes and subcloned into a pIZ/V5-His (Invitrogen, Thermo Fisher Scientific, Inc., USA). The pIZ/V5-His containing NA, namely pIZ-NA, was later transfected into Sf9 insect cells.
Insect cell Transfection and Transformation
pIZ-NA was transfected into Sf9 insect cells with the assistance of Cellfectin® II reagent (Invitrogen, Thermo Fisher Scientific, Inc., USA) according to the manufacturer's protocol. Briefly, pIZ-NA and Cellfectin® II reagent were mixed in Grace’s insect medium without serum supplement and incubated with freshly seeded Sf9 insect cells. After incubation of transfected cells at 27 °C for 3–5 h, the transfection mixture was removed and replaced with Grace’s insect medium supplemented with 10% Fetal Bovine Serum (FBS). Seventy-two hours post-transfection, NA gene expression in the transfected cells was determined by PCR of reverse transcription as described in gene expression analysis below, while the recombinant NA protein produced and secreted into the transfected culture medium was detected by Western blot. Transfected cells, which had been shown to express the NA gene and produce recombinant NA protein, were then split into a 1:5 ratio with the fresh medium and cells were left to attach to the tissue culture flask before adding the selective medium, the modified Graces’s insect medium containing 300 μg/ml Zeocin™ (Invitrogen, Thermo Fisher Scientific, Inc., USA). This selective medium was replaced every 3 to 4 days until resistant cells formed colonies to obtain stably transformed polyclonal populations. These stably transformed cells were allowed to grow to confluence and cloned by dilution method described below for isolation and selection of stable transformant cell lines.
Cloning of Stably Transformed Sf9 Cell Lines
Briefly, 10 ml of 1 × 104 cells/ml of stably transformed cells in the modified Grace’s insect cell medium was prepared, and the cell suspension (100 µl/well) was added to a 32 wells of a 96-well plate. The remaining cell suspension was further diluted at 1:1 with the same medium, and a100 μl/well of this cell suspension was again added to the next 32 wells. A similar procedure was repeated with the last 32 wells. Cells were left to attach overnight, then the medium was replaced with the fresh medium containing Zeocin™ and incubated at 27 °C for 1 week. Wells that contained only one colony, implying that it had originated from a single cell, were marked, and incubated until the colony fills most of the wells. These isolated cells were then harvested as cloned cells, tested for the NA gene expression by reverse transcription-PCR and recombinant NA protein production by Western blot analysis. Selected clones were expanded by cultured in 12- and 6-well plates, and finally in a T-25 flask.
Gene Expression Analysis
NA gene expression analysis was carried out by PCR of reverse transcription (RT-PCR). The transfected cells were harvested and extracted by addition of TRIZOL (Invitrogen, USA) and reverse transcription was carried out using oligo (dT) primers following the manufacturer’s instruction (Thermo scientific, USA) for cDNA synthesis. The synthesized total cDNAs were then used as templates for PCR and primers specific to NA sequences at 5ʹ and 3ʹ; forward primer: 5ʹ-GGGGTACCAAAATGAATCCAAATAAG-3ʹ and reverse primer: 5ʹ-GTGAATTCCTACTTGTCAATGGTG-3ʹ were used for detection of a full-length NA transcript.
SDS/ PAGE and Western Blot
Culture supernatant of transfected Sf9 cells was collected and prepared in the presence or absence of 2% β-mercaptoethanol before being subjected to 10% SDS-PAGE and Western blot for detection of recombinant NA protein. Proteins on SDS-PAGE were visualized by Coomassie staining (InstantBlue®, Abcam, USA) or silver staining (PlusOne silver staining kit, GE Healthcare, Sweden). For recombinant NA protein detection by Western blot, proteins from the SDS/PAGE were transferred to the nitrocellulose membrane. The membrane was next incubated in mouse anti-NA monoclonal antibody (H1N1) (Sino Biological, China) diluted at a 1:5 dilution ratio in 1% Bovine Serum Albumin (BSA) in PBST (PBS with 0.1% Tween™ 20) and left at 4 °C overnight and goat-anti-Mouse IgG (Abcam, China) conjugated with horseradish peroxidase diluted at a 1: 5000 ratio in 1% Bovine Serum Albumin (BSA) in PBST for 2 h at room temperature. The signal on immunoblot was developed with TMB conjugate substrate kit (Sigma-Aldrich, USA). NA-VLPs yields were estimated by comparison of the NA-specific band intensities on the Western blot, determined by an image analysis program (ImageJ), with the NA-specific band of the purified, known concentration, NA-VLPs. For rapid NA-VLPs detection, a dot-blot assay using the same procedure as the Western blot analysis was also performed, except that protein samples were directly dotted onto the nitrocellulose membrane.
Electron Microscope Analysis
Culture medium from the stably transformed Sf9 clonal cell culture was loaded onto 30% sucrose cushion and ultracentrifuged at 145,400×g for 2.5 h at 4 °C in a Sorvall WX100 + Ultracentrifuge (Thermo scientific, USA). The pellet was resuspended in PBS and applied onto a formvar carbon coated grid (EMS, USA) and left for 10 min. After washing in water, the grid was stained with 2% (v/w) phosphotungstic acid pH 7.2 (Sigma, USA) for 10 min and air dried. VLPs were observed using Hitachi HT7700 TEM microscope at 100 kV. Immuno-gold labeling method was also performed. A formvar carbon coated grid was floated on the surface of a drop of sample suspension and left at room temperature for 30 min. The grid was next floated onto an anti-NA monoclonal antibody drop diluted at 1:500 ratio in PBS for 1 h. After three times washing with PBS, the grid was floated onto an anti-mouse IgG conjugated with 5 nm gold particle drop (Sigma, USA) diluted at 1:2000 ratios in PBS for 30 min. After three times washing with PBS, the grid was stained with 2% phosphotungstic acid (pH 7.2) for 10 min. The air-dried grid was examined by a transmission electron microscope HT7700 (Hitachi, Japan).
Purification of NA-VLPs by Sucrose Density Gradient Ultracentrifugation
Stably transformed Sf9 cells culture medium containing the NA-VLPs was clarified by centrifuge at 5500 rpm for 20 min then the supernatant was layered onto a 30% sucrose cushion and centrifuged at 145,400×g for 2.5 h at 4 °C in a Sorvall WX100 + Ultracentrifuge (Thermo scientific, USA). The pellet was resuspended in PBS and loaded onto a sucrose gradient (20–60%) and centrifuged at 209,600×g for 2 h at 4 °C in the Sorvall WX 100 + Ultracentrifuge (Thermo scientific, USA). The sucrose layers from 40 to 50% were drawn and re-centrifuged at 145,400×g for 2.5 h at 4 °C. The purified NA-VLPs in the pellet were resuspended in PBS and filtered sterilized with 0.2 μm filtration and stored at − 20 °C.
Immunization
All immunizations were performed at the Department of Medical science, Ministry of public Health, Thailand, with approval from the ethics committee for animal experimentation (project number 61-034). The NA-VLPs purified by the sucrose gradient ultracentrifugation as previously described was used for immunization. Three of five weeks old female BALB/c mice were intramuscularly injected with 5 μg of NA-VLPs mixed with 2% Alhydrogel adjuvant (InvivoGen, USA) at a ratio of 1:1 (v/v). Two negative controls were a mouse injected with Phosphate Buffer Saline (PBS) and a mouse injected with 15 μg of a purified recombinant Japanese encephalitis virus envelope protein (JEV714), a kind gift from Asst. Prof. Dr. Anan Tongta (School of Bioresources and Technology, KMUTT, Thailand) which had been mixed with the same adjuvant as previously described. A booster was administered to each mouse 3 weeks after the first immunization. All mice were sacrificed 9 weeks after the primary immunization and sera collected. Specific antibody raised by immunized antigens in mice sera were determined by Western blot.
Neuraminidase Inhibition (NAI) Assays
Sera collected from sacrificed mice were diluted in PBS and NAI assays were performed according to a modified and miniaturized Warren’s thiobarbituric acid (TBA) method by Sandbulte et al. [28]. Diluted serum (10 µl) was mixed with equal volume of influenza quadrivalent inactivated split vaccine (FluQuadri®, Sanofi Pasteur, India) containing 2 influenza A subtypes (H1N1 and H3N2) and 2 influenza B (Victoria lineage and Yamagata lineage) then incubated for 30–45 min at room temperature. Fetuin solution (25 mg⁄ml) 10 µl was added to mixtures of serum and vaccine and incubated at 37 °C for 15–16 h. Four fetuin control tubes were prepared by adding fetuin solution into 20 µl of PBS in replace of serum and vaccine mixtures. Next, 10 µl periodate reagent (200 mM NaIO4 in 53% H3PO4), was added and incubated at room temperature for 15–20 min. Arsenite reagent (1 M AsNaO2, 700 mM Na2SO4, 0.03% concentrated H2SO4) 50 µl was added, and agitated until the yellow color disappeared. Thiobarbituric acid reagent (50 mM TBA, 625 mM Na2SO4) 10 µl was then added into each tube, incubated for 15 min at 99 °C on a dry bath then cooled down on ice. Warrenoff reagent (95% 1-butanol and 5% HCl) was dispensed at 150 µl to each tube and vortexed vigorously until the pink chromophore in the organic layer was observed. The mixer was centrifuge at 250×g for 5 min and 50 µl of the upper phase was transferred into a 96-well half flat bottom plates and absorbances at 550 nm (Thermo Scientific, USA) were measured. The absorbance obtained in fetuin control wells was subtracted from readings of all other wells as mean background. Assay results were accepted if the mean OD550 values of virus control samples (no serum) were 0.45–0.85 and fetuin control samples were < 0.1. The serum dilution that corresponded to a value of 50% in the following equation is defined as the NAI50 titer of the tested serum: OD value of tested serum/OD value of control serum × 100 (%). The NAI titer was defined as the dilution of the test serum at the initial point of the titration, which finally provided the NAI50. All assays were performed in triplicate.
Results
Stably Transformed Sf9 Insect Cells Producing Multimeric Recombinant NA-Proteins
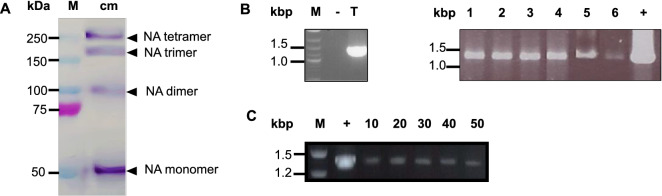
The full-length NA gene was subcloned into a pIZ/V5-His, namely pIZ-NA, and later transfected into Sf9 insect cells. The presence of Zeocin™ resistant gene in this plasmid enabled the transfected cells to withstand the Zeocin™ in the medium. These cells also produced and released multiple proteins into the culture medium which were recognized by anti-NA antibody as shown by Western blot (Fig. 1A). Their apparent sizes in the non-reducing condition were approximately 55 kDa, 110 kDa, 165 kDa, and 220 kDa. According to the NA size and structure, they could be monomer, dimer, trimer, and tetramer of the NA protein, respectively. To further confirm that these proteins were products of the NA gene expression, the transfected cells were collected and their total RNAs were subjected to gene expression analysis by PCR of reverse transcription method. A PCR product at the same size of the NA gene transcripts was observed (Fig. 1B Lane T) indicating that these transfected cells expressed the NA gene. As some cells might not be transfected with the plasmid, cell cloning using dilution method was carried out to obtain monoclones for transfected cell isolation. PCR of reverse transcription was performed for screening of clonal cells those are able to express the NA gene. Six clones were selected as a specific NA transcript was amplified in each sample (Fig. 1B) and the recombinant NA proteins were also detected in their culture medium by dot-blot (Fig. S1, supplementation). Clone no. 2 was randomly chosen as a representative for further studies, including analysis of stability of the NA gene expression. After 50 consecutive passages, NA gene expression of clone no. 2 was still detected (Fig. 1C). Other clones also had similar NA gene expression stability profiles (data not shown). Thus, these clonal cells were stably transformed Sf9 cells capable of expressing NA gene and produce recombinant NA protein.
Fig. 1.
A Western blot of culture medium (cm) obtained from the pIZ-NA transfected Sf9 cells, prepared in non-reducing condition, using anti-NA monoclonal antibody. Lane M: marker proteins. B PCR of reverse transcription for NA gene expression analysis of transfected cells (T) and clone cells no.1–6 of the pIZ-NA transfected Sf9 cells. C PCR of reverse transcription for NA gene expression analysis of clone no.2 at passage no. 10, 20, 30, 40, and 50. − : negative control for PCR using non-transfected cells cDNA, + : positive control for PCR using pIZ-NA as a template
NA-VLPs Formation
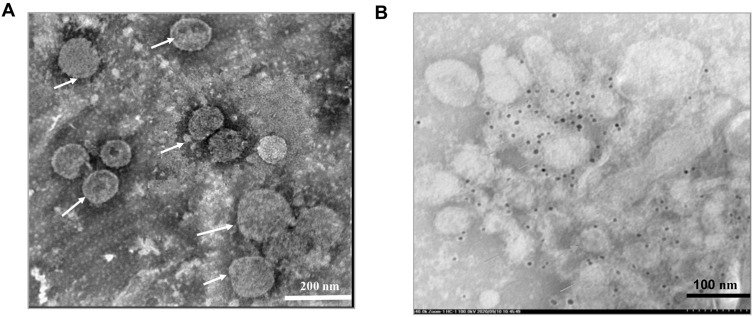
To investigate whether recombinant NA proteins at various sizes found on the Western blot (Fig. 1A) could assemble into virus-like particles, electron microscopic analysis of the concentrated culture medium was carried out. Electron micrographs of negatively stained sample on the grid showed large, spherical, enveloped virus-like particles with sizes ranging from 80 to 150 nm in diameter (Fig. 2A). These particles were assembled forms of recombinant NA protein since they were specifically bound by anti-NA antibodies as illustrated by the immunogold labeling assay (Fig. 2B). Thus, the stably transformed Sf cell lines constructed in this study were able to produce virus-like particles from recombinant NA protein, namely NA-VLPs, and released them into the culture medium.
Fig. 2.
Electron micrograph of NA-VLPs from concentrated culture medium of the pIZ-NA transfected Sf9 cells. A Samples were negatively stained with 2% phosphotungstic acid. B The same samples labeled by anti-NA antibody followed by anti-mouse IgG conjugated with 5 nm gold. Arrows indicate NA-VLPs
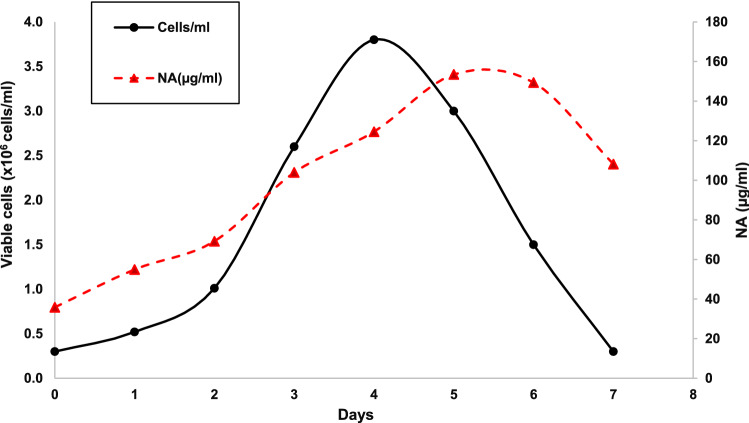
Stably Transformed Sf9 Insect Cell Growth and NA-VLPs Production
Growth of the stably transformed Sf9 cell (clone no.2), as suspension cells, in Grace’s insect medium supplemented with 10% FBS was found to be relatively similar to the non-transformed SF9 cells. They reached maximum cell density at 3.8 × 106 cells/ml on day 4 before entering the decline phase (Fig. 3). The NA-VLPs production was found to be associated with the cell growth and its level reached maximum, at approximately 150 µg/ml, on day 5. Since the Sf9 cells are known to be able to grow in the serum-free medium, these stably transformed Sf9 cell clonal cell lines were also adapted to be cultured in Sf900-II serum-free medium. These cells could also produce recombinant NA protein and form multimers (Fig.S2, Supplementation). However, many degraded NA proteins were observed even though the freshly harvested sample had been used.
Fig. 3.
Growth of stably transformed Sf9 cells, clone no. 2, in Grace’s complete medium containing 10% FBS and kinetics of NA-VLP production and secretion into the culture medium
NA-VLPs Purification
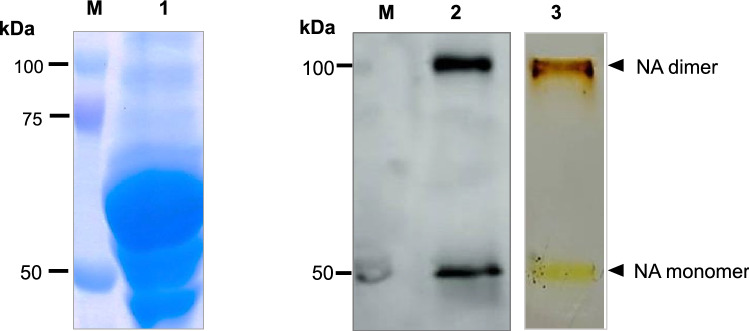
The NA-VLPs were originally produced and secreted by the stably transformed cells into the Graces’s medium supplemented with 10% FBS. The serum albumin which could be seen on the SDS/PAGE at approximately 58 kDa constitutes a great proportion of the total proteins in the medium (Fig. 4). Due to differences in sizes and weight of NA-VLPs and the serum albumin, a sucrose gradients ultracentrifugation was performed for NA-VLPs purification. After 30% sucrose cushion ultracentrifugation, the resuspended pellet was loaded onto 20–60% sucrose gradients. It was found that in the 40–50%sucrose layers, pure NA-VLPs were obtained (Fig. 4).
Fig. 4.
Recombinant NA protein in stably transformed Sf9 culture medium before and after a sucrose gradient ultracentrifugation. Lane 1: Coomassie-stained SDS/PAGE of culture medium before purification, Lane 2 and Lane 3: Western blot and silver-stained SDS/PAGE, respectively of the sample after the sucrose density gradient ultracentrifugation, from 40 to 50%sucrose layers, Lane M: standard protein markers
NA-VLPs Immunogenicity
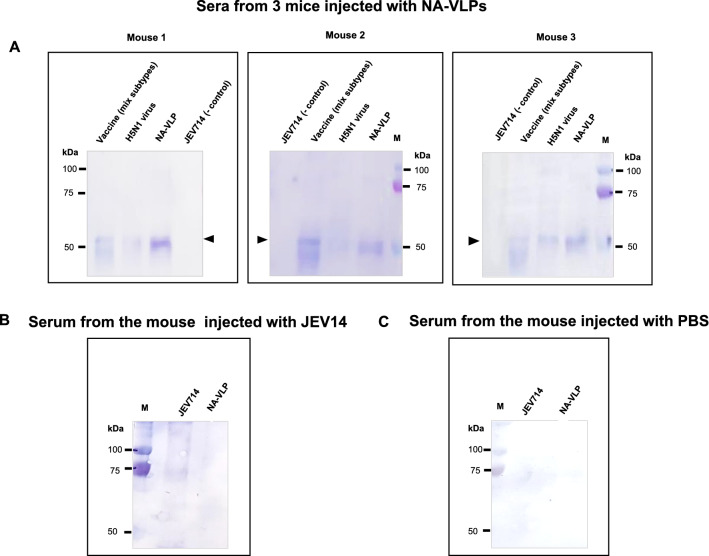
After the first injection and a booster with the NA-VLPs and controls, sera from immunized mice were collected and used as a primary antibody against NA-VLPs produced from stably transformed cells. These sera were also tested for their reaction to a mixture of commercial Influenza quadrivalent vaccine (FluQuardi®) and inactivated proteins from influenza A virus (H5N1) as positive controls and a non-related protein, a recombinant Japanese encephalitis virus envelope protein (JEV714), as a negative control. The results showed that sera from all 3 mice immunized with the NA-VLPs contained antibodies that could bind to the NA monomer, the 55 kDa protein. When these sera were tested against the FluQuardi® inactivated split influenza vaccine which contained two influenza A subtypes, H1N1 and H2N3, or inactivated proteins from influenza A virus (H5N1), a band of similar size of the recombinant NA protein was detected (Fig. 5A). These sera did not cross-react with the non-related recombinant Japanese Encephalitis envelope protein (JEV714). For the mouse that was immunized with the JEV714, its serum had an antibody that could bind to a protein in the JEV714 sample but not to the NA-VLPs (Fig. 5B). As expected, the serum from the PBS injected mouse contained no antibody that could bind to neither the NA-VLPs nor the JEV714 (Fig. 5C).
Fig. 5.
Detection of anti-NA antibodies in sera of mice immunized with 3 antigens, A NA-VLPs, B Recombinant Japanese encephalitis virus envelope protein (JEV714), non-related protein negative control and C Phosphate Buffer Saline, negative control. Protein samples on the Western blot were influenza quadrivalent vaccine (FluQuardi), H5N1: inactivated proteins from influenza A virus (H5N1), NA-VLPs and JEV714, M: standard protein marker. Triangles indicate NA monomer of NA-VLPs
NAI Activity
Sera collected from NA-VLPs immunized mice that had previously been shown to have anti-NA-VLPs antibodies were tested for their abilities to inhibit the neuraminidase activity of the influenza virus. In this study, the neuraminidase used for the NAI assay was from two influenza A, H1N1 and H3N2 and two strains of influenza B viruses in the influenza quadrivalent vaccine (FluQuardi®). Serial diluted sera, 1:1, 1:10, 1:40, and 1:160 were individually mixed with the vaccines and the neuraminidase inhibition activities (NAI) were determined. NAI titers were antilog10 of sera dilution that could inhibit 50% of NA activity. Table 1 showed different NAI titers of three NA-VLPs immunized mice sera. These results indicate that the NA-VLPs efficiently induce anti-NA antibody production in immunized mice and these antibodies efficiently inhibit the neuraminidase activity of the mixed subtypes of influenza viruses in the tested vaccines.
Table 1.
NAI titers of sera from 3 mice immunized with the NA-VLPs
| Mouse No. | 1 | 2 | 3 |
|---|---|---|---|
| NAI titer | 100 ± 5 | 79.4 ± 5 | 125.9 ± 10 |
Discussion
An established lepidopteran, Spodoptera frugiperda insect cell lines (Sf9) were genetically engineered by a plasmid-driven gene expression method to exclusively express the influenza NA gene and produce NA-VLPs. An expression plasmid containing the NA gene, pIZ-NA, was first constructed and then transfected into SF9 cells. Western blot of culture medium collected from the pIZ-NA transfected Sf9 cell shows bands recognized by anti-NA antibody at sizes that are corresponded to NA monomer, dimer, trimer, and tetramer (Fig. 1A). These results indicated a successful NA gene expression and production of a recombinant NA protein. Oligomerization of the recombinant NA protein detected on the Western blot are in agree with previous studies stating that NA oligomerization happens through an N-terminal directed process within the endoplasmic reticulum [29–31]. Thus, detection of multimeric recombinant NA proteins in the transfected cell culture medium were as expected. Zeocin™ was applied into the transfected cell culture until resistant polyclonal population established. This heterogeneous pool of cells had a range of recombinant NA protein productivities, depending on the NA gene copy number and site of chromosome integration. Cell cloning was therefore carried out to isolate and select only high-producer clones, to ensure maximum recombinant NA protein yields. Six clonal lines had been isolated, expanded, and their abilities to express the NA gene were confirmed by RT-PCR (Fig. 1B). All clones produced recombinant NA protein at comparable levels as determined by dot blot analysis (Fig. S1, Supplementation). A clone was randomly chosen for further studies. The selected clonal cell lines had been sub-cultured in the medium in the presences of Zeocin™ until their growth was stable, then this drug was no longer added. Even though without Zeocin™ in the culture, the NA gene expression could be detected for up to 50 passages (Fig. 1C). Their ability to maintain NA gene expression in the drug-free conditions has made them ideal cells for vaccine production.
As reported by Lai et al. [25], human embryonic kidney (HEK-293 T) cells, exclusively produced recombinant NA protein, were capable of efficiently forming VLPs which were morphologically similar to influenza virions [25]. In this study, Sf9 transformed cells expressing only the NA gene also revealed their abilities to synthesize, form NA-VLPs and release them into the medium. Electron micrographs of the concentrated culture medium showed particles with morphology similar to influenza virions. These particles were recognized by anti-NA antibody as shown by the binding of gold particles which in turn specifically bound to the anti-NA antibody in the immunogold assay. This results indicates that the virus-like particles were made from the recombinant NA proteins. (Fig. 2).
The stably transformed cells continuously secreted multimers of recombinant NA protein into the culture medium and the protein yield reached its maximum level on day 5, at approximately 150 µg/ml, and then declined when cells entered the dead phase (Fig. 3). This amount of recombinant NA protein produced was in the high range of the protein produced by insect cells due to the use of a baculovirus strong constitutive promoter, (OpMNPV) IE-2 [27, 32]. Constitutive NA gene expression allowed the accumulation of the recombinant NA protein as its level continued rising until day 6 when cells entered the dead phase (Fig. 3). Due to the absence of cell lysis of this system, the stably transformed Sf9 cell lines provided a stable physiological environment for the facilitation of NA assembly, translocation to the membrane, and secretion to the culture medium. Although these cell lines had been cultured in a serum-supplemented medium while being genetically engineered and cloned, they were able to adapt to grow in a serum-free medium, Sf900-II, and maintain their capability to produce NA-VLPs (Fig. S2, Supplementation). It is worth noting that the NA-VLPs from cells grown in the Sf900-II serum free medium were more rapidly degraded compared to the NA-VLPs from the serum-supplement medium. This could be due to the action of proteolytic enzymes which are generally secreted from cells into the medium. In the absence of serum, the NA-VLPs are the main target of these enzymes [33]. Proteolytic inhibitors are thus highly recommended when using serum-free medium.
The NA-VLPs could be directly purified from the culture medium of stably transformed cells by the sucrose gradient ultracentrifugation. Due to the size difference, the serum albumin was on the top layers of the sucrose gradients while only NA-VLPs were found between 40 and 50%. Sucrose layers. In the case of cells cultured in the serum-free medium, a simpler purification process such as ultrafiltration, for buffer exchange and removing some proteins released from cells [33] may be applied. It was suggested by many reports that the protection induced by the NA protein was mediated by NAI antibodies [34–38]. Thus, a goal for the development of the influenza NA-based vaccine is its ability to induce NAI antibodies for effectiveness in conferring protection against influenza viruses [39]. In this study, the NA-VLPs could very well stimulate NA-specific antibodies in immunized mice. Sera obtained from all mice were able to inhibit neuraminidase activities of quadrivalent vaccine (FluQuardi®) containing two influenza A, H1N1 and H3N2, and two strains of influenza B viruses as shown by their NAIs in Table 1. This was as anticipated, since VLPs present high-density NA-epitopes for antibody production. Thus, NA-VLPs are potential NA-based vaccine antigens for widening the breadth of protection against various influenza virus strains.
Conclusion
A simple process for NA-based influenza vaccine production by the stably transformed Sf9 insect cells had been developed. The NA antigen was made as virus-like particles solely from the recombinant NA protein without the presence of other influenza virus proteins. Using a plasmid-driven NA gene expression followed by cell cloning and selection, clonal stably transformed cell lines capable of supporting the NA protein post-translation and assembly into VLPs, were obtained. The NA-VLPs are efficiently trafficked to the plasma membrane and released into the culture medium. The constitutive expression of the NA gene in these cells leads to continuous production and secretion of NA-VLPs into the culture medium. This allows consistent and reproducible production of the target antigen as well as a simple process for purification of the NA-VLPs from the medium, i.e., a sucrose gradient ultracentrifugation. Both upstream and downstream processes had been designed to facilitate the scale-up for GLP/GMP production. This study provides a simple and high potential approach for encouraging the use of NA antigen in current influenza vaccines besides the largely HA-focused approaches.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Professor Pilaipan Puthavathana, Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand for kindly providing Influenza A virus (A/Thailand/1(KAN1) /2004/H5N1) neuraminidase gene and Asst. Prof. Dr. Anan Tongta (School of Bioresources and Technology, KMUTT, Thailand) for allowing us to join animal facilities and service at the Department of Medical science, Ministry of Public Health, Thailand and a kind gift, the purified recombinant Japanese encephalitis virus envelope protein (JEV714), used as a control in immunogenicity studies. Ms. Najmeh Khanefard was supported by Petchra Pra Jom Klao Ph.D. scholarship from King Mongkut’s University of Technology Thonburi, Thailand.
Funding
This research is funded by National Research Council of Thailand, Grant No. 107412.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gravel C, Li C, Wang J, Hashem AM, Jaentschke B, Xu KW, Lorbetskie B, Gingras G, Aubin Y, Van Domselaar G, Girard M, He R, Li X. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine. 2010;28:5774–5784. doi: 10.1016/j.vaccine.2010.06.075. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martinez-Alonso M, Fodor E. Conserved and host-specific features of influenza virion architecture. Nature Communications. 2014;5:4816. doi: 10.1038/ncomms5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ko EJ, Lee YT, Kim KH, Lee Y, Jung YJ, Kim MC, Lee YN, Kang T, Kang SM. Roles of aluminum hydroxide and monophosphoryl lipid a adjuvants in overcoming CD4+ T cell deficiency to induce isotype-switched IgG antibody responses and protection by T-dependent influenza vaccine. The Journal of Immunology. 2017;198:279–291. doi: 10.4049/jimmunol.1600173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tumpey TM, Renshaw M, Clements JD, Katz JM. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. Journal of Virology. 2001;75:5141–5150. doi: 10.1128/JVI.75.11.5141-5150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo M. Influenza virus entry. Advances in Experimental Medicine and Biology. 2012;726:201–221. doi: 10.1007/978-1-4614-0980-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlanda SF, Tsvetnitsky V, Donnelly JJ. Universal influenza vaccines: Shifting to better vaccines. Vaccine. 2016;34:2926–2933. doi: 10.1016/j.vaccine.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nature Reviews Drug Discovery. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 8.Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. Journal of Virology. 1993;67(6):2972–2980. doi: 10.1128/JVI.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichelberger MC, Wan H. Influenza neuraminidase as a vaccine antigen. Current Topics in Microbiology and Immunology. 2015;386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 10.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. Journal of Virology. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. Journal of Biological Chemistry. 2010;285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laver WG, Valentine RC. Morphology of the isolated hemagglutinin and neuraminidase subunits of influenza virus. Virology. 1969;38(1):105–119. doi: 10.1016/0042-6822(69)90132-9. [DOI] [PubMed] [Google Scholar]
- 13.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feele EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du R, Cui Q, Rong L. Competitive cooperation of hemagglutinin and neuraminidase during influenza A virus entry. Viruses. 2019 doi: 10.3390/v11050458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAuley JL, Gilbertson BP, Trifkovic S, Brown LE, McKimm-Breschkin JL. Influenza virus neuraminidase structure and functions. Frontiers in Microbiology. 2019;10:39. doi: 10.3389/fmicb.2019.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen F, Wan XF. Influenza neuraminidase: Underrated role in receptor binding. Trends in Microbiology. 2019;27(6):477–479. doi: 10.1016/j.tim.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilbourne ED, Laver WG, Schulman JL, Webster RG. Antiviral activity of antiserum specific for an influenza virus neuraminidase. Journal of Virology. 1968;2:281–288. doi: 10.1128/jvi.2.4.281-288.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. Journal of Infectious Diseases. 1974;129:411–420. doi: 10.1093/infdis/129.4.411.Couchetal.1974. [DOI] [PubMed] [Google Scholar]
- 19.Murphy BR, Kasel JA, Chanock RM. Association of serum anti neuraminidase antibody with resistance to influenza in man. New England Journal of Medicine. 1972;286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- 20.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristo lT, Fargis S, Risos K, Powers JH, Davey RT, Taubenberger JK. Evaluation of anti-hemagglutinin and anti-neuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio. 2016;19(7):e00417–16. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon M, Kirkpatrick E, Stadlbauer D, Strohmeier S, Bouvier NM, Krammer F. Mucosal immunity against neuraminidase prevents influenza B virus transmission in guinea pigs. mBio. 2019;10:e00560–19. doi: 10.1128/mBio.00560-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wohlbold TJ, Krammer F. In the shadow of hemagglutinin: A growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses. 2014;6:2465–2494. doi: 10.3390/v6062465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krammer F, García-Sastre A, Palese P. Is it possible to develop a “universal” influenza virus vaccine? Potential target antigens and critical aspects for a universal influenza vaccine. Cold Spring Harbor perspectives in biology. 2018;10(7):a028845. doi: 10.1101/cshperspect.a028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sridhar S, Brokstad KA, Cox RJ. Influenza vaccination strategies: Comparing inactivated and live attenuated influenza vaccines. Vaccines. 2015;3(2):373–389. doi: 10.3390/vaccines3020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai JC, Chan WW, Kien F, Nicholls JM, Peiris JS, Garcia JM. Formation of virus-like particles from human cell lines exclusively expressing influenza neuraminidase. Journal of General Virology. 2010;91(Pt 9):2322–2330. doi: 10.1099/vir.0.019935-0. [DOI] [PubMed] [Google Scholar]
- 26.Harrison RL, Jarvis DL. Transforming lepidopteran insect cells for continuous recombinant protein expression vol. 1350. In: Murhammer D, editor. Baculovirus and Insect Cell Expression Protocols Methods in Molecular Biology. New York: Humana Press; 2016. pp. 329–348. [DOI] [PubMed] [Google Scholar]
- 27.Theilmann DA, Stewart S. Molecular analysis of the trans-activating IE-2 gene of Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology. 1992;187:84–96. doi: 10.1016/0042-6822(92)90297-3. [DOI] [PubMed] [Google Scholar]
- 28.Sandbulte MR, Gao J, Straight TM, Eichelberger MC. A miniaturized assay for influenza neuraminidase-inhibiting antibodies utilizing reverse genetics-derived antigens. Influenza and Other Respiratory Viruses. 2009;3(5):233–240. doi: 10.1111/j.1750-2659.2009.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito T, Taylor G, Webster RG. Steps in maturation of influenza A virus neuraminidase. Journal of Virology. 1995;69:5011–5017. doi: 10.1128/jvi.69.8.5011-5017.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogue BG, Nayak DP. Synthesis and processing of the influenza virus neuraminidase, a type II transmembrane glycoprotein. Virology. 1992;188:510–517. doi: 10.1016/0042-6822(92)90505-J. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Glidden EJ, Murphy SR, Pearse BR, Hebert DN. The cotranslational maturation program for the type II membrane glycoprotein influenza neuraminidase. Journal of Biological Chemistry. 2008;283:33826–33837. doi: 10.1074/jbc.M806897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegedus DD, Pfeifer TA, Hendry J, Theilmann DA, Grigliatti TA. A series of broad host range shuttle vectors for constitutive and inducible expression of heterologous proteins in insect cells. Gene. 1998;207:241–249. doi: 10.1016/S0378-1119(97)00636-7. [DOI] [PubMed] [Google Scholar]
- 33.Shin J, Rhim J, Kwon Y, Choi SY, Shin S, Ha C, Lee C. Comparative analysis of differentially secreted proteins in serum-free and serum-containing media by using BONCAT and pulsed SILAC. Science and Reports. 2019;9:3096. doi: 10.1038/s41598-019-39650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, Belmont JW. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. Journal of Infectious Diseases. 2013;207:974–981. doi: 10.1093/infdis/jis935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT, Jr, Taubenberger JK. Evaluation of anti-hemagglutinin and anti-neuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. mBio. 2016;7:e00417–16. doi: 10.1128/mBio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stadlbauer D, Zhu X, McMahon M, Turner JS, Wohlbold TJ, Schmitz AJ, et al. Broadly protective human antibodies that target the active site of influenza virus neuraminidase. Science. 2019;366:499–504. doi: 10.1126/science.aay0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel OA, Manicassamy B. Broadly protective strategies against influenza viruses: Universal vaccines and therapeutics. Frontiers in Microbiology. 2020;11:135. doi: 10.3389/fmicb.2020.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X, Wang Q, Liu M, Zheng Q, Wu F, Huang J. Tetrameric neuraminidase of influenza a virus is required to induce protective antibody responses in mice. Frontiers in Microbiology. 2021;12:729914. doi: 10.3389/fmicb.2021.729914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim K, Lee Y, Park S, Jung Y, Lee Y, Ko E, Kim Y, Li X, Kang S. Neuraminidase expressing virus-like particle vaccine provides effective cross protection against influenza virus. Virology. 2019;535:179–188. doi: 10.1016/j.virol.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.