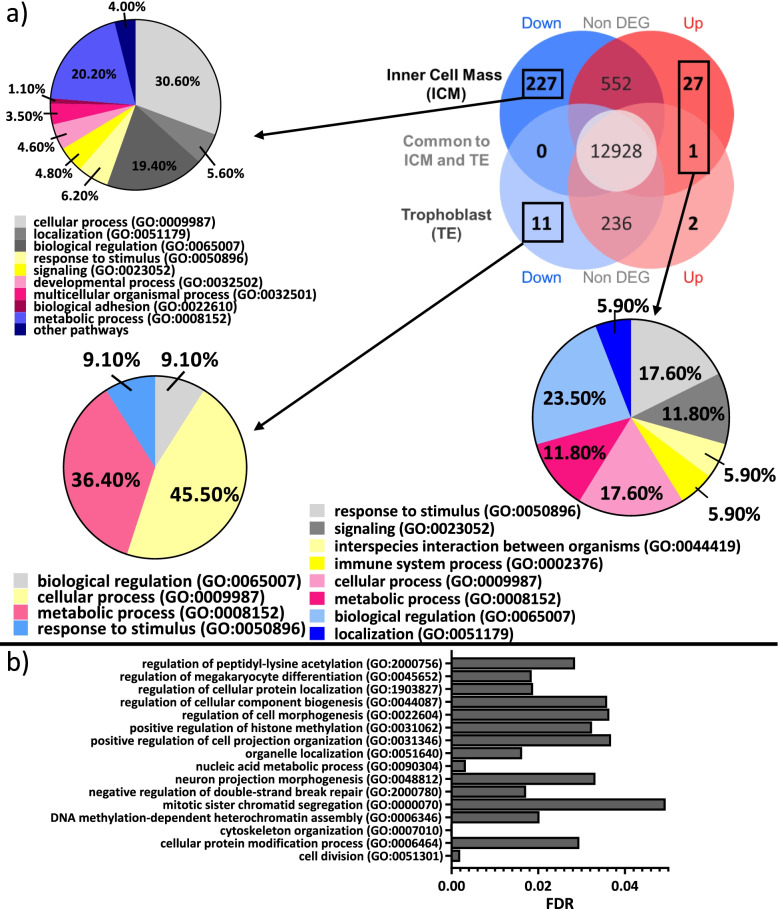
Fig. 3.
Analysis of differentially expressed genes (DEG) in embryos according to maternal age. A representation of down (blue) and upregulated (red) DEG in ICM (from DeMixT_ICM_cells data obtained after deconvolution of ICMandTE using DeMixT R package [38, 39]) and TE (from TE_part dataset) of embryos from OM. Nota bene: YM is the reference group. Functional classifications (for down regulated genes in ICM and TE and for up-regulated genes in ICM) using GO Biological Process obtained with PANTHER online software are presented as pie charts. For upregulated DEG in TE, no functional classification was made. B Bar chart presenting p-adjusted of most specific subclass pathways provided by PANTHER online software after statistical overrepresentation test (Fisher’s Exact test and Benjamini-Hochberg correction) with the Human GO Biological Process annotation on down-regulated DEGs in ICM of OM. Nota bene: YM is the reference group. DEG: Differentially Expressed Genes (FDR < 0.05); TE: Trophoblast; ICM: Inner Cell Mass; OM: Old mares; YM: young mares