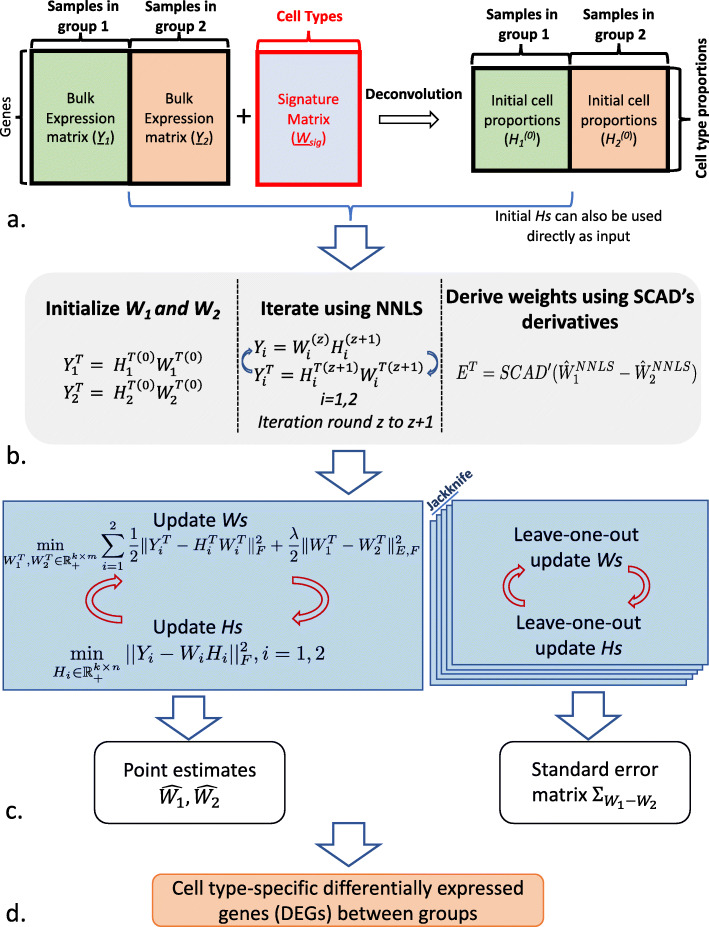
Fig. 1.
Schematic diagram for SCADIE: (a) SCADIE requires either (1) bulk gene expression matrices and cell type proportions or (2) bulk gene expression matrices and shared signature matrix as input; the cell type proportions can be obtained by any deconvolution method. (b) The initial full W matrices and weights are obtained by a few rounds of NNLS-based iterative updates. (c) The main iterative procedure consists of two parts: one iterative run for estimating , and n (sample size) additional runs for leave-one-out jackknife estimation for standard error . (d) With point estimates and entry-wise standard errors, DEGs can thus be identified