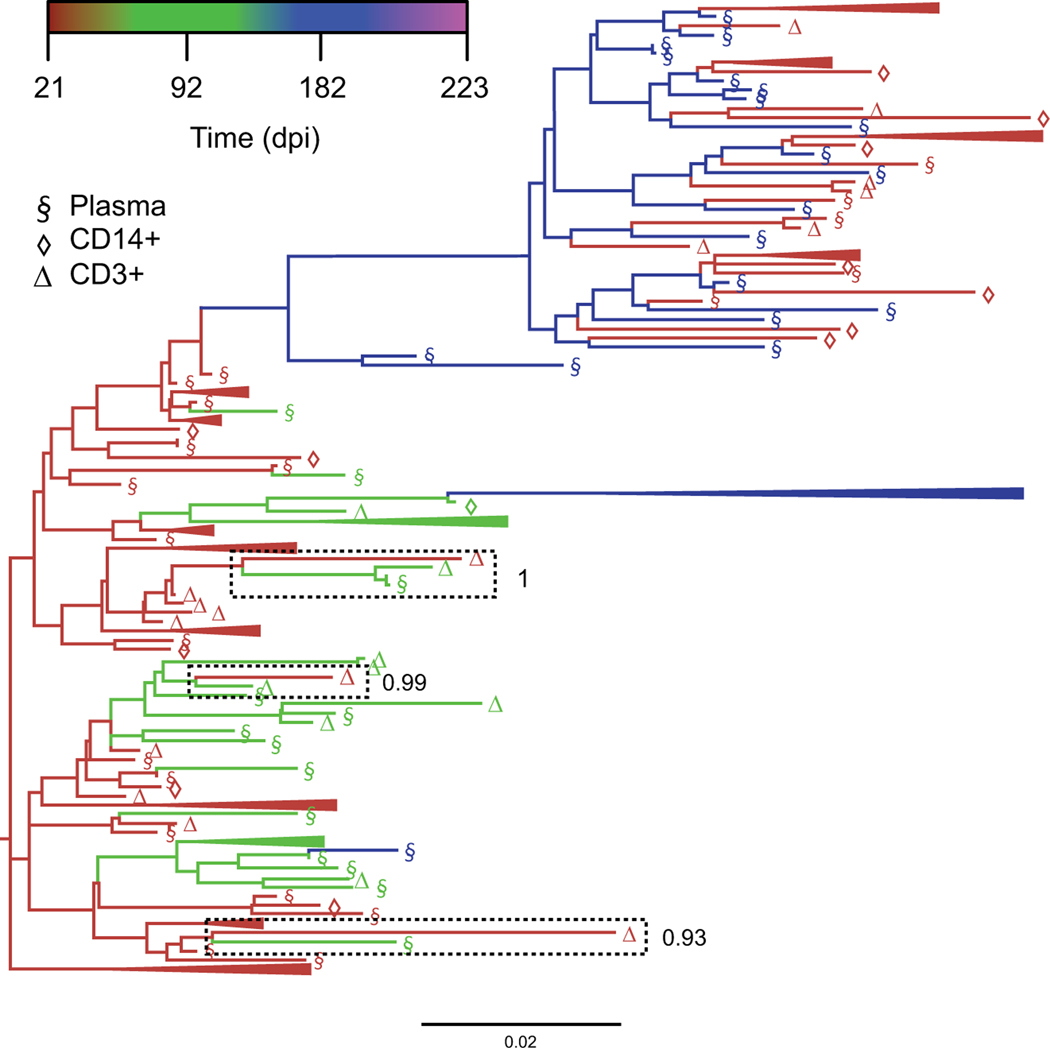
Fig. (2). Bayesian maximum clade credibility (MCC) tree highlighting reduced intra-host temporal structure.
The coalescent framework implemented in BEAST [111, 112] was used for a serially sampled SIV-infected macaque dataset to obtain a posterior distribution of trees scaled in substitutions/site, from which the MCC tree was derived using Tree Annotator [112]. Branches are colored according to sampling time point in days post-infection (dpi), with temporally clustered sequences (>3) collapsed for illustrative purposes. Three separate instances of shared ancestry between sequences sampled relatively far apart (92 and 223 dpi) have been highlighted, with corresponding clade posterior probabilities reported to the right for which the temporal states have been randomized. The final result is referred to as the temporal clustering statistic, ranging from 0 (absence of temporal structure) and 1 (perfect temporal structure).