Abstract
In 2004, the identification of female germline or oogonial stem cells (OSCs) that can support post–natal oogenesis in ovaries of adult mice sparked a major paradigm shift in reproductive biology. Although these findings have been independently verified, and further extended to include identification of OSCs in adult ovaries of many species ranging from pigs and cows to non–human primates and humans, a recent study rooted in single–cell RNA sequence analysis (scRNA-seq) of adult human ovarian cortical tissue claimed that OSCs do not exist, and that other groups working with OSCs following isolation by magnetic-assisted or fluorescence-activated cell sorting have mistaken perivascular cells (PVCs) for germ cells. Here we report that rare germ lineage cells with a gene expression profile matched to OSCs but distinct from that of other cells, including oocytes and PVCs, can be identified in adult human ovarian cortical tissue by scRNA-seq after optimization of analytical workflow parameters. Deeper cell-by-cell expression profiling also uncovered evidence of germ cells undergoing meiosis-I in adult human ovaries. Lastly, we show that, if not properly controlled for, PVCs can be inadvertently isolated during flow cytometry protocols designed to sort OSCs because of inherently high cellular autofluorescence. However, human PVCs and human germ cells segregate into distinct clusters following scRNA-seq due to non–overlapping gene expression profiles, which would preclude the mistaken identification and use of PVCs as OSCs during functional characterization studies.
Keywords: human ovary, single, cell RNA sequence analysis, oocyte, germ cell, oogonial stem cell
Graphical Abstract
Graphical Abstract.
Significance Statement.
The recent discovery of a rare stem cell population in the ovaries of women that is capable of supporting the production of new eggs cells or oocytes has the potential to significantly change the current landscape for the clinical management of female infertility as well as the hormonal imbalance resulting from ovarian failure at menopause. This study further documents the existence of these rare cells in the ovaries of women, the genetic profile of these cells, and the occurrence of the earliest steps of the differentiation of these cells into new oocytes in the ovaries of women under normal physiological conditions.
Introduction
A central underpinning of reproductive biology has held that oocyte generation in ovaries of female mammals is restricted to the embryonic period.1 This thinking deviates markedly from spermatogenesis in males throughout adult life, which involves meiotic differentiation of male germline or spermatogonial stem cells (SSCs) in the testes.2 However, the longstanding paradigm of a non–renewing oocyte pool was challenged by a study with mice in 2004, which reported the existence of female germline or oogonial stem cells (OSCs) and the continuation of oocyte production in adult mouse ovaries.3 While this study sparked significant debate,4,5 more than 80 corroborating studies now support the existence of OSCs and/or active oogenesis in adulthood across species,6 including humans7-15 (Supplementary Tables S1–S3). The discovery of OSCs, which brings the biology of male and female gametogenesis in mammals more closely in line with one another and with that of non-vertebrate species,16 has significant ramifications for the development of in vitro models to investigate human oogenesis as well as of new technologies to combat ovarian failure and female infertility caused by aging or insults.17-19
A major breakthrough in the study of OSCs came in 2009, with the first report that the cells could be retrieved as a distinct population from mouse ovaries using DEAD-box helicase 4 (DDX4) antibody-based sorting.20 Through extensive in vitro characterization and in vivo transplantation studies, the germline identity of the cells was established, as was the functional identity of the cells as bona fide precursors to oocytes that can be fertilized to produce viable offspring.20 More than 60 other publications have since isolated OSCs from ovaries of mice, rats, pigs, cows, baboons, and humans6 (Supplementary Tables S1 and S2). Moreover, inducible genetic lineage tracing studies with mice have fate–mapped new oocytes produced during adulthood to the generation of healthy offspring in natural mating trials, thus establishing the physiological importance of OSC-supported oogenesis to adult ovarian function and female fertility.21 A second major breakthrough came in 2012 with the successful purification of OSCs from adult human ovarian cortical tissue,7,8 the findings of which have since been independently verified and extended by many other groups.9,11,12,14,15 Human OSCs express a profile of genes characteristic of primitive germ cells, and these cells differentiate through meiosis into oocyte–like cells in vitro and into oocytes that are enclosed within newly formed follicles after transplantation into human ovarian cortical tissue.7-15,17 Human OSCs isolated by fluorescence-activated cell sorting (FACS) with monoclonal antibodies against DDX4 have also been used in approved clinical studies.17,22-24
Discordant with this large body of work, a recent study concluded from single–cell RNA sequence analysis (scRNA–seq) that human OSCs do not exist.25 These authors identified only 6 clusters (viz. 6 cell types) in adult human ovarian cortical tissue biopsies using scRNA-seq: stromal cells, perivascular cells (PVCs), endothelial cells, granulosa cells, immune cells, and oocytes. That non–oocyte germ lineage cells were apparently missing from their analysis was subsequently put forth as evidence of OSCs being absent from adult human ovaries. However, the expression and clustering analysis reported in this study were performed with an early version of Cell Ranger software (2.1.1 or v2), which has widely known limitations in its ability to detect low-expression cells. An improved version of Cell Ranger software (3.0.2 or v3), which was available and actually used by the authors in the same study for analysis of human ovarian cells after flow cytometric sorting, increases the sensitivity for cell calling by approximately one log-order over that using Cell Ranger v226 (https://www.10xgenomics.com). While the preprocessed data obtained from Cell Ranger v2 and v3 are fairly consistent, the ability of Cell Ranger v3 to detect more cells, especially those with low abundance transcripts, can change batch-effect corrections and thus the accuracy of the output data analysis.26 Another issue that can affect the resolution of scRNA-seq is the human reference genome used for read alignments, with HG38 preferred over HG19 for optimal depth of analysis.27 All of this is highly relevant because OSCs, like other stem cell types, are very rare, with a reported frequency of ~0.014% in adult ovaries.7 If one is seeking to identify as many cells, and as many cell types, as possible in a highly heterogenous cell sample using scRNA-seq, decisions about which analytical approaches will be used become critically important to consider prior to performing the experiments.26,27
In this same study, Wagner et al.25 also used DDX4 antibody-based FACS coupled with scRNA-seq to claim that OSCs isolated and studied by many others for over a decade using the same cell sorting strategy7-13, 20, 22, 23, 28 (see also14, 15Supplementary Table S1) are actually PVCs lacking any germ lineage characteristics. Given the past debate over whether mammalian OSCs exist,4-6 and with our labs being directly involved in studies of OSCs for nearly 2 decades,3,7,8,10,11,13,19,29,30 we felt it was important to experimentally assess the conclusions reached by Wagner et al.25 in an effort to reconcile this recent report with the opposite conclusions reached in over 60 other published studies that have isolated and characterized OSCs since 20096 (Supplementary Tables S1 and S2). In parallel, we evaluated the possibility that technology like scRNA-seq could provide further evidence of not just the existence of human OSCs but also of primitive germ cells committing to, or progressing through, the early stages of meiotic differentiation into oocytes in adult human ovarian tissue under normal physiological conditions in vivo. Such an outcome, which has not yet been demonstrated in humans, would be consistent with recent genetic tracing studies in adult female mice showing that active meiotic entry and oogenesis occur naturally in adult ovaries during reproductive life,21,31,32 and that oocytes formed in the ovaries during adulthood contribute directly to the pool of eggs used for natural species propagation.21
Materials and Methods
Animals
Freshly collected ovarian tissues from adult heifers (Bos taurus) were obtained from Blood Farms (Groton, MA) and processed immediately for cortex isolation and cryopreservation until use.
Human Subjects
All research with human tissues was approved by the institutional review boards of Northeastern University (IRB#14-03-22), University of Edinburgh (LREC 16/SS/0144), and Saitama Medical University (630-III). Informed consent was obtained from all participants, and all tissue samples were de-identified prior to use. A total of 7 ovarian cortical tissue samples from 2 caesarean section patients (CSP) and 1 gender reassignment patient (GRP) between 26 and 34 years of age7,8,11 were used.
Adult Human Unsorted Ovarian Cortical Cell scRNA-seq Data and Code Availability
The scRNA-seq data referenced in this study were originally generated by Wagner et al.25 from adult human ovarian cortical biopsies of 4 subjects (CSP, n = 3; GRP, n = 1). The 10× Genomics dataset of Wagner et al.25 for adult human unsorted ovarian cortical cells was deposited by these authors to, and accessed by us through, the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) under the accession code E-MTAb−8381. Analyses of scRNA-seq data were completed using the lines of code for adult human unsorted ovarian cortical cells deposited by Wagner et al.25 on GitHub (https://github.com/wagmag/SingleCellOvary). For Cell Ranger v6 analysis, additional lines of code were run in parallel to select proper quality control metrics as well as to determine the parameters for dimensionality reduction that best represented the data. The code used with the Cell Ranger v6 analysis is available on GitHub (https://github.com/hanrico/Ovarian-scRNA-seq).
Clustering and Analysis of scRNA-seq Data
In the Wagner et al.25 study, output files for their adult human unsorted ovarian cortical cell samples were converted using Cell Ranger v2. We first re-analyzed their raw fastq files using the same version of Cell Ranger and the same human genome assembly (HG19), along with Seurat version 3.0.0 (v3) and the lines of code for unsorted human ovarian cortical cells deposited by Wagner et al.25 We then repeated this analysis using Cell Ranger version v3, since this newer software version was also available to the authors at the time of their study. In fact, Wagner et al.25 elected to use Cell Ranger v3 for their analysis of sorted ovarian cells, but for unclear reasons, they chose Cell Ranger v2 for their unsorted ovarian cell analysis. Finally, the same dataset was analyzed using current versions of Cell Ranger (version 6.0.1 or v6) and Seurat (version 4.0.4 or v4), along with HG38 as the human reference genome (Supplementary Table S4). After completing the Seurat analysis of the Cell Ranger v6 output data, we then replaced Seurat with Loupe Browser (version 5.1.0 or v5; 10× Genomics) for deeper expression analysis of single cells, using the same filtering and data visualization parameters utilized with Seurat (see Supplementary material, Method 1 for additional details).
Expression of PRDM1 (PR domain containing 1 with ZNF domain), DPPA3 (developmental pluripotency-associated 3), IFITM3 (interferon-induced transmembrane protein 3), TUBB8 (tubulin beta 8 class VIII), and DDX4 was used to identify primitive germ cells, noting that Wagner et al.25 used a more limited profile of only PRDM1, DPPA3, and DAZL (deleted in azoospermia like). Analysis of FIGLA (factor in the germline alpha), OOSP2 (oocyte secreted protein 2), GDF9 (growth differentiation factor 9), and ZP3 (zona pellucida glycoprotein 3) was used to identify oocytes, as reported by Wagner et al.25 However, we also analyzed the expression of ZP1, ZP2, and NOBOX (newborn ovary homeobox) as oocyte markers. Expression of SYCP3 (synaptonemal complex protein 3), STAG3 (stromal antigen 3), SMC1a (structural maintenance of chromosomes 1 alpha), SMC3 (structural maintenance of chromosomes 3), and STRA8 (stimulated by retinoic acid gene 8) was used to identify germ cells in the first meiotic division. Expression of RGS5 (regulator of G-protein signaling 5), MCAM (melanoma cell adhesion molecule), MYH11 (myosin heavy chain 11), RERGL (Ras-related and estrogen-regulated growth inhibitor-like), and TAGLN (transgelin) was used to identify PVCs, as reported by Wagner et al.25 For ease of referral, a listing and brief overview of each gene analyzed is provided in Supplementary Table S5.
Flow Cytometry
Ovarian cortical tissue from adult heifers or reproductive-age women was cryopreserved, thawed, and dissociated into single–cell suspensions for flow cytometry using a BD FACSAria™ III, as described previously8,33 (see Supplementary material, Method 2 for more details). Primary antibodies against SMA (ab5694, 1:50; Abcam, Cambridge, MA) or CD31 (MA3100, 1:50; Invitrogen–ThermoFisher Scientific, Waltham, MA), each directly conjugated to APC (Abcam, ab201807), were used for determination of the total percentage of autofluorescent events that were positive for expression of either PVC marker. Flow cytometry data were acquired using BD FACSDiva 8.0.1 software and analyzed by FlowJo 10.7 software.
Data Analysis
For scRNA-seq, data analysis was performed using the human unsorted ovarian cortical cell dataset of Wagner et al.25 For FACS, the data represent the mean ± SE of combined results; n = 4 (CD31) or 7 (SMA), and n = 5 (SMA) or 6 (CD31), for bovine and human ovarian tissue sample analysis, respectively.
Results
Analysis of Unsorted Cells Isolated from Human Ovarian Cortical Tissue Biopsies
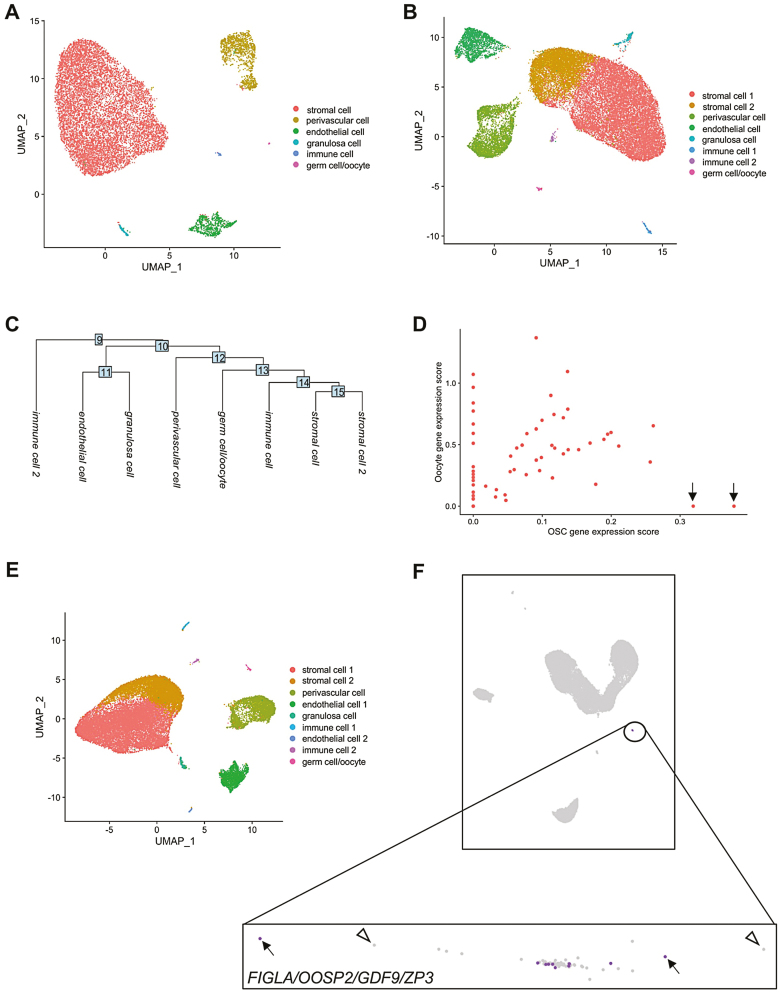
We first used the reported frequency of OSCs in adult ovarian tissue with software for estimating the number of cells required to detect a given cell type with scRNA-seq (www.satijalab.org/howmanycells). Assuming 6 general cell types based on the clusters reported by Wagner et al.,25 we determined that 84 550 viable ovarian cells would be needed for detection of at least 5 OSCs at 95% confidence (Supplementary Fig. S1). Any number less than 5 cells failed to produce a reliable assessment of input cell number required. Notably, the 12 160 cells analyzed by Wagner et al.25 was 14.4% of the minimal cell input number needed for detection of 5 OSCs at 95% confidence under these modeling parameters (see Supplementary material, Method 3 for additional details). To then assess if the use of Cell Ranger v2 versus v3 would have altered the outcomes reported by Wagner et al.,25 we reanalyzed their adult human unsorted ovarian cortical cell dataset using Cell Ranger v2 and v3 with their published code, HG19, and Seurat v3. With Cell Ranger v2, we identified the 6 clusters reported by these authors from 12 020 total cells called after preprocessing and filtering the Cell Ranger output data (Fig. 1A). When we switched to Cell Ranger v3, we identified the same 6 clusters plus 2 additional clusters that emerged from more than double the number (27 376) total cells called (Fig. 1B, 1C). Through gene ontology (GO) analysis, the 2 additional clusters were enriched for genes associated with stromal and immune cells. Wagner et al.25 reported that 1 of the 6 clusters they identified, which was comprised of 18 total cells, exhibited higher expression levels of 4 markers commonly associated with oocytes (FIGLA, OOSP2, GDF9, and ZP3) relative to expression levels of these genes in the other 5 clusters. Our analysis using Cell Ranger v2 similarly identified 18 cells that constituted a single cluster enriched for expression of these 4 genes (Supplementary Fig. S2; Supplementary Table S6). However, when analyzed using Cell Ranger v3, this cluster increased from 18 to 62 cells (Supplementary Table S6). A deeper analysis of this cluster identified 2 cells, not detected with Cell Ranger v2, that were positively associated with genes used by Wagner et al.25 to identify OSCs (PRDM1, DPPA3, and DAZL) but had an oocyte gene expression score of zero (Fig. 1D).
Figure 1.
Clustering and analysis of unsorted adult human ovarian cortical cells following scRNA-seq. (A) Identification of a total of 12 020 cells that formed 6 clusters following analysis of the dataset using Cell Ranger v2. (B) Identification of a total of 27 376 cells that formed 8 clusters following reassessment of the same dataset with Cell Ranger v3. (C) Cluster dendrogram depicting the lineage relationships between the 8 clusters identified using Cell Ranger v3. (D) Scatterplot analysis of the 62-cell germ cell/oocyte cluster identified using Cell Ranger v3, showing OSC gene expression scores plotted against oocyte gene expression scores. Two cells with very high OSC gene expression scores and an oocyte gene expression score of zero are highlighted by black arrows. (E) Identification of a total of 27 710 cells that formed 9 clusters following reassessment of the dataset with Cell Ranger v6. (F) Loupe Browser analysis of the Cell Ranger v6 output data, with the germ cell/oocyte cluster highlighted by the expanded box. Of the 62 cells in this cluster (each cell is depicted as an individual dot), 10 cells were identified as positive for co-expression of FIGLA, OOSP2, GDF9, and ZP3 (purple dots; examples are highlighted by black arrows) whereas 52 cells did not show co-expression of this 4-gene oocyte marker panel (light-gray dots; examples are highlighted by open arrowheads).
Workflow Adjustments to Optimize scRNA-seq of Human Ovarian Cortical Cells
Our identification of at least 2 potential non-oocyte germ cells using Cell Ranger v3 that were missed when Cell Ranger v2 was used prompted us to ask if further optimization of the scRNA-seq workflow could provide additional insights into whether evidence of OSCs in this dataset could be uncovered. To do this, we analyzed the same dataset using current versions of Cell Ranger software (v6) and Seurat (v4), along with HG38 and with the number of principal components, dimensions, and resolution set at 30, 1:18, and 0.1, respectively. Using this updated workflow, UMAP analysis identified 9 clusters from a total of 27 710 cells called (Fig. 1E; Supplementary Table S6). Using GO analysis, we identified the clusters as likely representing stromal cells (2 separate clusters), PVCs, endothelial cells (2 separate clusters), granulosa cells, immune cells (2 separate clusters), and germ cells/oocytes. We then compared outcomes obtained using Seurat v4 versus Loupe Browser v5 for downstream analysis of the preprocessed Cell Ranger v6 data, since Loupe Browser is more user-friendly and does not require specific lines of code, like Seurat, for interactive visualization of the results. Both Seurat and Loupe Browser identified the same germ cell/oocyte cluster based on similarity in overall expression patterns. However, cell-by-cell analysis with Loupe Browser revealed that only 10 of 62 total cells in this cluster co-expressed all 4 marker genes associated with oocytes (FIGLA, OOSP2, GDF9, and ZP3) (Fig. 1F). However, since premeiotic and post–meiotic germ cells are known to express a common suite of genes that define the germ lineage, we suspected that these 62 cells were likely clustered based on germline, rather than on oocyte-associated, gene expression patterns.
Analysis of Human Unsorted Ovarian Cortical Cells for Evidence of Oocytes
Using our optimized analytical pipeline, 10 cells were identified in the germ cell/oocyte cluster as oocytes based on co-expression FIGLA, OOSP2, GDF9, and ZP3 in each cell (Fig. 1F). While FIGLA is often referred to as an oocyte-specific gene,34-36FIGLA encodes a transcription factor that functions at various stages of oogenesis, including the regulation of key genes needed for meiosis-I progression in premeiotic germ cells.37 We, therefore, performed a gene-by-gene analysis of the 62 cells in this cluster using Loupe Browser. We identified a total of 40 FIGLA-expressing cells, of which only 21 co-expressed OOSP2, 13 co-expressed GDF9, 37 co-expressed ZP3, and 12 co-expressed both GDF9 and ZP3 (Fig. 2). Likewise, when the other oocyte markers were analyzed individually, we identified 25 OOSP2-expressing cells, 14 GDF9-expressing cells, and 42 ZP3-expressing cells in this cluster (Fig. 2). The discordance in numbers of cells expressing each gene individually or in pairs versus combined as a 4-gene panel may be due to differences in the timing of expression of the various genes relative to oocyte maturational stage, expression in cells other than oocytes, and/or degradation of mRNA transcripts during sample processing that led to the expression of a given gene in a given cell falling below the detection threshold (see Discussion and Supplementary material, Discussion 1 for additional details).
Figure 2.
Cell-by-cell analysis of oocyte-associated markers in the germ cell/oocyte cluster using Cell Ranger v6 followed by Loupe Browser. Cells identified as positive for expression of the indicated gene(s) are dark shaded in purple (examples are highlighted by black arrows in the uppermost panel) whereas cells negative for expression of the indicated gene(s) are light shaded in gray (examples are highlighted by white arrowheads in the uppermost panel).
While DDX4 is widely accepted as being expressed in all oocytes in vivo,7,38-40 only 12 of the 62 cells in this cluster expressed DDX4 (Fig. 3) Moreover, only 5 of the FIGLA/OOSP2/GDF9/ZP3-expressing cells co-expressed DDX4 (Fig. 3). Pairwise gene analysis identified 12 DDX4/FIGLA-expressing cells, 8 DDX4/OOSP2-expressing cells, 6 DDX4/GDF9-expressing cells, and 10 DDX4/ZP3-expressing cells (Fig. 3). Thus, out of the 4 marker genes used by Wagner et al.25 to identify oocytes, the only gene co-expressed in all DDX4-expressing cells in this cluster was FIGLA, which is expressed in both pre– and post–meiotic germ cells.37 A parallel analysis of NOBOX, which in mouse and human ovaries is robustly expressed in oocytes throughout development from the primordial follicle to metaphase-II egg stage,41,42 failed to identify a single NOBOX-expressing cell in the germ cell/oocyte cluster (Fig. 3). Likewise, there were no ZP1-expressing cells, and only 2 ZP2-expressing cells, identified in this cluster of cells containing candidate oocytes (Fig. 3). Of the 2 ZP2-expressing cells, only 1 co-expressed the 4-gene marker panel used by Wagner et al.25 to identify oocytes, whereas the other co-expressed FIGLA, OOSP2, and ZP3, but not GDF9 (data not shown).
Figure 3.
Further analysis of the germ cell/oocyte cluster for expression of DDX4 and oocyte markers on a cell-by-cell basis using Loupe Browser. Cells identified as positive for expression of the indicated gene(s) are dark shaded in purple whereas cells negative for of the indicated gene(s) are light shaded in gray. No ZP1-expressing cells were identified in this cluster (data not shown).
Analysis of Human Unsorted Ovarian Cortical Cells for Evidence of Non–Oocyte Germ Cells
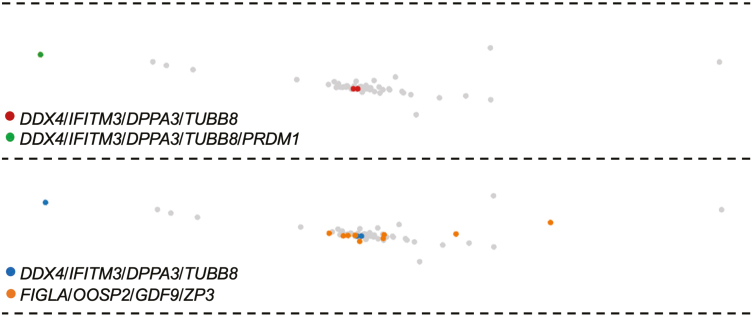
We next moved to the analysis of genes known to be expressed in primitive germ cells (see Supplementary Table S5 for more details). We identified 1 cell in the germ cell/oocyte cluster with the expression of all 5 germline genes analyzed (PRDM1, DPPA3, IFITM3, TUBB8, and DDX4), and 2 additional cells with expression of DPPA3, IFITM3, TUBB8, and DDX4 but lacking detectable PRDM1 (Fig. 4). All 3 cells localized to the same cluster of 62 cells which contained the 10 FIGLA/OOSP2/GDF9/ZP3-expressing cells. However, these 3 cells were clearly distinct from the 10 cells classified as oocytes (Fig. 4). In the 2 non–oocyte germ cells lacking PRDM1 expression, we detected expression of SYCP3 (Table 1), which is required for the progression of germ cells through the early stages of the first meiotic cell division.43,44 This observation prompted us to explore additional genes involved in the early stages of meiosis-I. From this, we found that both SYCP3-expressing germ cells co-expressed STAG3 and SMC3, and 1 of the SYCP3/STAG3/SMC3-expressing germ cells also co-expressed SMC1a (Table 1). Notably, the proteins encoded by STAG3, SMC1a, and SMC3 are all meiosis-specific cohesin complex components involved in the formation of axial elements and cohesion of sister chromatids during meiotic prophase-I.45-48 We also identified 2 SYCP3/STAG3/SMC3-expressing cells in this cluster with co-expression of STRA8 (Supplementary Fig. S3), the latter of which is considered a critical early gene for meiosis-I progression in germ cells of both sexes.49
Figure 4.
Loupe Browser analysis of the germ cell/oocyte cluster for expression of genes highly enriched in primitive germ cells. Cells identified as positive for expression of the indicated gene(s) is/are colorized (see legends in each panel) whereas cells negative for expression of the indicated gene(s) are light shaded in gray.
Table 1.
Gene expression profiling analysis of non–oocyte germ cells identified in adult human ovarian cortical tissue. Loupe Browser analysis of the 3 non–oocyte germ lineage cells for expression of oocyte marker genes, genes associated with meiosis-I activation and progression, and PVC marker genes (−, expression not detected; +, expression detected).
| DDX4/IFITIM3/DPPA3/ TUBB8 Cell 1 | DDX4/IFITIM3/DPPA3/ TUBB8 Cell 2 | DDX4/IFITIM3/DPPA3/TUBB8/PRDM1 | |
|---|---|---|---|
|
FIGLA/OOSP2/
GDF9/ZP3 |
− | – | – |
| FIGLA | + | + | + |
| OOSP2 | + | + | − |
| GDF9 | − | – | − |
| ZP3 | + | + | + |
| SYCP3 | + | + | − |
| STAG3 | + | + | − |
| SMC1a | + | − | − |
| SMC3 | + | − | − |
|
RGS5/MCAM/
MYH11/RERGL/ TAGLN |
− | − | − |
| RGS5 | − | − | − |
| MCAM | − | − | − |
| MYH11 | − | − | − |
| RERGL | − | − | − |
| TAGLN | + | + | + |
It is worth noting that FIGLA was detected in all 3 DPPA3/IFITM3/TUBB8/DDX4-expressing cells (Table 1). However, since these cells were distinct from the FIGLA/OOSP2/GDF9/ZP3-expressing cells (viz. candidate oocytes), the presence of FIGLA, which is not oocyte-specific,37 is still aligned with these 3 cells being classified as non–oocyte germ cells. Expression of OOSP2 was detected in the 2 DPPA3/IFITM3/TUBB8/DDX4-expressing cells, but not in the single PRDM1/DPPA3/IFITM3/TUBB8/DDX4-expressing cell (Table 1). While OOSP gene family members were first identified as encoding oocyte-enriched proteins in the mouse ovary,50,51 lineage specificity of OOSP2 in human ovaries has not been evaluated to date, and transcriptomic expression of the gene in humans is not restricted to oocytes.52
Continuing with our analysis, GDF9 was not detected in any of the DPPA3/IFITM3/TUBB8/DDX4-expressing cells found in this cluster, whereas ZP3 was detected in all 3 cells (Table 1). However, ZP3 expression was far more ubiquitous than expected, in that a total of 567 cells with ZP3 expression were identified across the population of 27 710 cells called in this dataset (Supplementary Fig. S4). Strikingly, 525 of these ZP3-expressing cells were localized outside of the germ cell/oocyte cluster (see Supplementary material, Results 1 for additional details). This widespread detection of ZP3 expression across clusters representing different lineages, most of which are somatic, is consistent with the reported low-level expression of this gene in diverse cell types and tissues in humans.52 To further assess the promiscuous nature of ZP3 expression outside of oocytes, we analyzed a different scRNA-seq dataset derived from adult human ovarian medullary tissue.53 We did not identify a single cell with co-expression of the 4-gene marker profile used by Wagner et al.25 to identify oocytes; however, parallel analysis of this dataset identified 673 cells expressing ZP3, again distributed randomly across the various clusters (data not shown).
Finally, given that Wagner et al.25 used DAZL as 1 of their 3 genes for OSC screening, we identified 20 DAZL-expressing cells in the entire dataset of 27 710 cells called using Cell Ranger v6, 10 of which were localized to the germ cell/oocyte cluster. Of these 10 cells, 5 co-expressed the 4-gene marker panel used by Wagner et al.25 to identify oocytes (Supplementary Fig. S5), consistent with past studies establishing expression and function of DAZL in both pre– and post–meiotic germ cells.54,55 Breaking the oocyte marker panel down further, we identified 8 DAZL/FIGLA-expressing cells, 8 DAZL/OOSP2-expressing cells, 8 DAZL/FIGLA/OOSP2-expressing cells, 5 DAZL/GDF9-expressing cells, 5 DAZL/ZP3-expressing cells, and 5 DAZL/GDF9/ZP3-expressing cells in the germ cell/oocyte cluster (data now shown).
Analysis of Cells Sorted from Human Ovarian Cortical Tissue Biopsies using Flow Cytometry
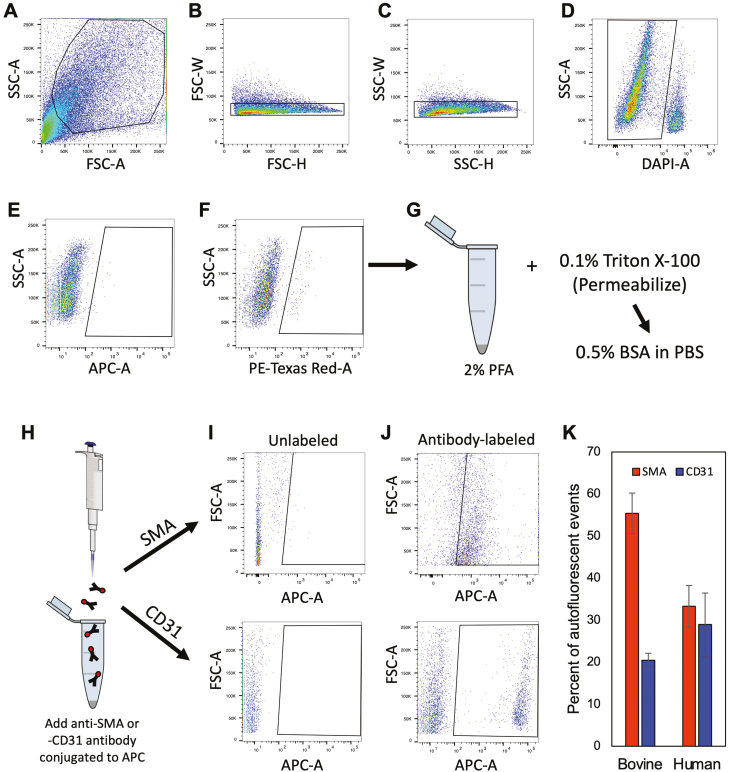
In the Wagner et al.25 study, the authors also reported that DDX4 antibody-based FACS, a method used by many others to specifically sort OSCs across species since 20097-13,20,22,23,28-30 (Supplementary Table S1), led to the isolation of PVCs and not OSCs. While initially puzzling, we noted that their flow cytometry was performed using AF594 detected with a 561-nm laser in the red channel, which is widely known to detect autofluorescence as a “positive” event during FACS. In evaluating the antibody validation and gating strategies shown in the Supplementary data of Wagner et al.25, we observed that the area above the cutoff designating the negative versus positive fractions in their negative control sample lacking antibody contained positive events, which represent autofluorescence. With this information in mind, PVCs are known to express autofluorescent biomolecules, such as collagen and elastin, which produce widely known artifacts in flow cytometry.56,57 We therefore sorted dispersed ovarian cortical tissue with a 561-nm laser in the red channel following the parameters published by Wagner et al.25 Using adult bovine ovarian cortical tissue for validation, a distinct population of autofluorescent events was obtained (Fig. 5). Three-quarters of these cells were positive for SMA or CD31, which respectively mark the 2 cell types that comprise PVCs: vascular smooth muscle cells and pericytes. A distinct population of autofluorescent events was similarly detected in dispersed ovarian cortical tissue of reproductive-age women. Likewise, almost two-thirds of these events were identified as being SMA- or CD31-positive (Fig. 5). Moreover, these autofluorescent events were detectable in dispersed cell preparations from human ovarian cortical tissue irrespective of whether the samples were gated versus FSC-A or SSC-A, or if PE-Texas Red-A was plotted against a different laser (APC-A) (Supplementary Fig. S6).
Figure 5.
Flow cytometric detection, isolation, and characterization of autofluorescent events in adult cow and human ovarian cortical tissue. (A–D) Representative gating strategy for doublet discrimination (forward-scatter or FSC-A: B; side-scatter or SSC-A: C) and for dead cell exclusion using 4ʹ,6-diamidino-2-phenylindole (DAPI) labeling (D). (E, F) Comparison of autofluorescent events detected in the APC-A far-red channel (640-nm laser; E) versus the PE-Texas red-A channel (561-nm laser; F). (G–K): Autofluorescent events detected in the PE-Texas red-A channel were collected, fixed and permeabilized (G and H), and then incubated with APC-conjugated primary antibodies against SMA (Abcam ab5694) or CD31 (Invitrogen MA3100) (I and J) for determination of the total percentage of autofluorescent events that were positive for expression of either PVC marker in bovine and human ovarian cortical tissue samples (K). Data shown in (K) are the mean ±SE; n = 4 (CD31) or 7 (SMA), and n = 5 (SMA) or 6 (CD31), for bovine and human sample analysis, respectively.
Germ Cells and PVCs Segregate into Distinct Clusters
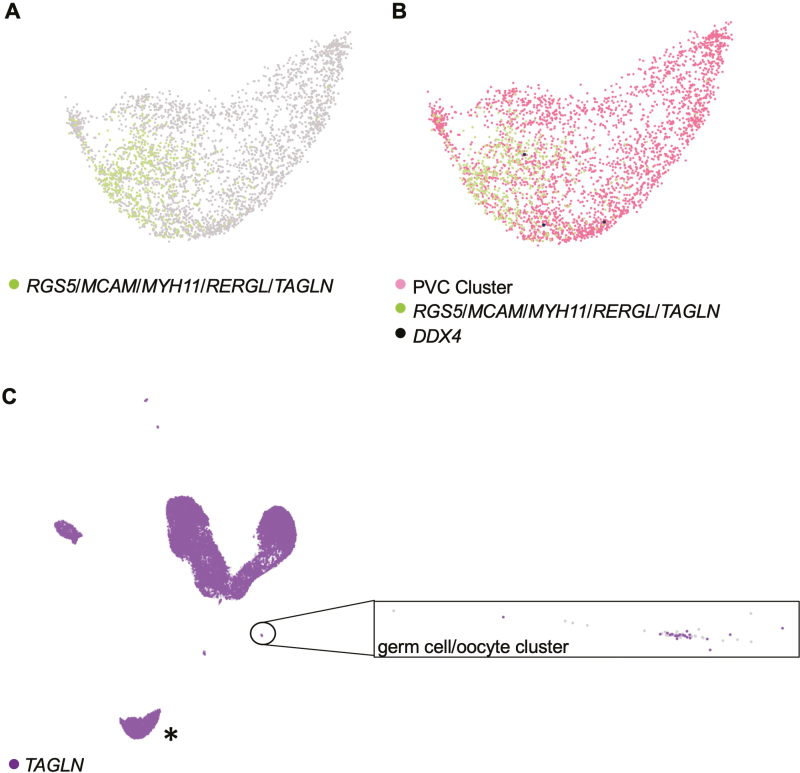
We then dug deeper into the conclusion of Wagner et al.25 that we and others have mistakenly worked with PVCs in studies that have explored the characteristic germline properties, and potential clinical utility, of human OSCs7-15,17,22,23 (Supplementary Tables S1 and S2). Using the optimized scRNA-seq workflow described above, we identified a cluster comprised of 3310 total cells, 479 of which had the 5-gene gene expression profile used by Wagner et al.25 to identify PVCs (RGS5, MCAM, MYH11, RERGL, and TAGLN) (Fig. 6A). None of the cells comprising this cluster co-expressed the gene panel which identified the 3 non-oocyte germ cells (DPPA3, IFITM3, TUBB8, and DDX4, without or with PRDM1; data not shown), and only 3 of the 3310 total cells in this cluster expressed DDX4 (Fig. 6B). Moreover, none of the 479 cells identified as PVCs using the 5-gene profile of Wagner et al.25 co-expressed DDX4 (Fig. 6B). Likewise, none of the 3 non-oocyte germ cells expressed the 5-gene profile used by Wagner et al.25 to cluster PVCs (Table 1), which led to the expected segregation of these 2 cell types into distinct and non-overlapping clusters (Fig. 6C). In fact, of the 5 genes used by Wagner et al.25 to cluster PVCs, TAGLN was the only gene identified through individual gene-by-gene analysis to be co-expressed in the 3 DPPA3/IFITM3/TUBB8/DDX4-expressing germ cells (Table 1). However, TAGLN expression is not specific to any single cell type, and thus its utility as a lineage marker must be viewed in context with other genes as a profile associated with a given cell type. Supporting this statement, we found that 16 291 cells of the 27 710 total cells called expressed TAGLN (Fig. 6C). This included 23 cells in the 62-cell germ cell/oocyte cluster, 4 of which co-expressed the 4-gene profile used by Wagner et al.25 to identify oocytes (Fig. 6C).
Figure 6.
Analysis of PVC markers in adult human unsorted ovarian cortical cells using Loupe Browser. (A) Identification of cells with co-expression of RGS5, MCAM, MYH11, RERGL, and TAGLN (green dots) in a larger population of cells clustered together as PVCs based on overall similarities in gene expression. (B) Assessment of DDX4 expression (black dots) in the PVC cluster, highlighting the absence of DDX4 in the subpopulation of cells that co-express RGS5, MCAM, MYH11, RERGL, and TAGLN (green dots). (C) Analysis of TAGLN expression (purple dots) across all cells called in the adult human unsorted ovarian cortical scRNA-seq dataset, with an expanded view of the germ cell/oocyte cluster shown (cells lacking expression of TAGLN are shown as light gray). Note that the cluster highlighted by the asterisk in (C) is the PVC cluster shown in (A) and (B). Light-gray dots: cells negative for expression of the indicated gene(s). See also related data shown in Table 1.
Discussion
While scRNA-seq is useful as a tool to gain insights into cell lineage heterogeneity within a sample,58 a major caveat of this approach is that its failure to detect gene expression-based evidence of a given cell type after clustering analysis does not, by default, equate to that cell type being absent in the sample analyzed. This is especially apropos in attempts to identify either very rare cells or low-expression cells in a dispersed cell preparation that is heterogenous in nature, highlighting the challenges associated with the detection of stem cells in tissues by scRNA-seq.59 The analytical pipeline used also has a significant impact on the depth and accuracy of the data obtained, especially if the objective is to produce a comprehensive snapshot of as many cells, and as many cell types, as possible in the sample analyzed. In the Wagner et al.25 study, their attempts to identify OSCs in a pool of 12 160 total cells called, given the extreme rarity of OSCs in adult ovaries,7 would be difficult even under the best of conditions (see Supplementary material Discussion 1 for additional details). Our rigorous reassessment of their unsorted cell dataset following empirical testing of numerous variables that affect the depth, resolution, and accuracy of scRNA-seq highlight how multiple decisions made by these authors for their analysis of unsorted cells actually minimized, rather than optimized, the probability of detecting rare or low-expression cell types such as OSCs. In fact, several other ovarian stem cell types were also missed by Wagner et al.,25 including pluripotent embryonic stem cell (ESC)-like cells, mesenchymal stem cells (MSCs), and very small embryonic-like stem cells (VSELs) (Supplementary Table S7; see also Supplementary material Discussion 2), the latter of which have been reported to support postnatal oogenesis in mammalian ovaries.32
These challenges were further complicated by the fact that Wagner et al.25 restricted their efforts to find evidence of OSCs in their entire dataset of 12 180 cells to only 15 cells that were manually selected by these authors based on the required presence of DDX4 mRNA to establish the only “cluster” of cells that could contain OSCs. The a priori assumption that all candidate OSCs must have detectable DDX4 expression using scRNA-seq is fraught with interpretational problems. For example, we found that only 5 of the 10 cells identified as oocytes using the 4-gene profile reported by Wagner et al.25 co-expressed DDX4. Based on their reasoning, such an approach would have removed the remaining 5 FIGLA/OOSP2/GDF9/ZP3-expressing cells lacking detectable DDX4 mRNA from further consideration as oocytes. Likewise, we could not identify a single NOBOX–expressing cell in the germ cell/oocyte cluster, even though NOBOX is highly expressed in oocytes at all developmental stages.41,42 If one evaluated this dataset for evidence of oocytes based solely on NOBOX expression, or manually created a “cluster” of NOBOX–expressing cells as the sole cell population in which any potential oocytes would be found, the reasoning of Wagner et al.25 with OSCs would lead to the erroneous conclusion that oocytes do not exist in adult human ovarian cortex. At the other end of the spectrum, our evaluation of ZP3, which is widely used as an “oocyte-specific” marker,35 revealed low-level but widespread expression of this gene across all cell clusters, most of which are somatic in origin. Thus, scRNA-seq workflow design, and any conclusions drawn, based on the expression of a single gene being detected or not in a cell of interest lack scientific rigor and confidence.
However, optimization of the scRNA-seq workflow pipeline using a version of Cell Ranger (v3), which was available to, and used by, Wagner et al. at the time of their study25 but was not used for their unsorted cell analysis, allowed us to identify rare cells in their adult human unsorted ovarian cortical cell dataset with a gene expression profile that closely aligns with that of primitive germ cells, such as embryonic PGCs60-63 and adult ovary-derived OSCs.6-9,11,12,20 Further analysis showed that 2 of these non–oocyte germ cells expressed multiple markers of meiosis-I commitment and progression. These observations, which offer evidence of ongoing de-novo oogenesis in adult human ovaries under normal physiological conditions, are consistent with prior studies with mice which have demonstrated that resident germ cells routinely undergo meiosis in adult ovaries.21,31,32 Interpretational caution must still be exercised here, however, since gene expression profiling does not offer definitive evidence of the existence, or not, of human OSCs or of active meiotic entry. In this regard, all scientific studies of isolated human OSCs published to date have characterized the cells, following isolation, by gene expression profiling along with various downstream functional tests of meiotic cell division capability and/or oocyte-forming potential.7-15 Like other gene expression-based studies, the inability of scRNA-seq as a standalone approach to offer any type of functional verification of suspected lineage identity is another caveat of the use of this type of “big-data” technology and the interpretations drawn from it.64
Moving on to the FACS analysis of cells identified by Wagner et al.25 as DDX4-positive (+) or DDX4-negative (−), the sorting strategy for OSC isolation relies on the detection of an externalized region of the C terminus of DDX4 exposed on the outside of viable cells and not simply DDX4 expression.7,8,20,28 Dual-antigen single-protein sorting studies conducted almost 10 years ago showed that OSCs could be sorted as viable cells using C-terminal, but not N-terminal, DDX4 antibodies, noting that both antibodies recognize DDX4 in oocytes in fixed ovarian sections.7 In turn, the viable cells sorted with the C-terminal antibody show a near-complete population shift by FACS when the same cells are permeabilized and then analyzed with an N-terminal DDX4 antibody,7 verifying the specificity of the sorting protocol for detection of externalized DDX4. It has also been shown that proper tissue dispersion is a crucial step to achieve viable cell isolation and to release OSCs as single cells.8,20,28 Human ovary tissue is particularly fibrous and difficult to disaggregate, and thus extreme care must be exercised during disaggregation to maintain cell viability.8,11,20,28 If this is not done, the possibility of non-specific antibody binding is markedly increased, which may explain why the yield of “DDX4+” events obtained by Wagner et al.25 with the Abcam DDX4 antibody was 3.0-6.5–fold higher than the yield of human OSCs reportedly previously using the same sorting approach with the same antibody.7
Putting potential technical issues aside, it is important to emphasize that no other study that has used DDX4 antibody–based sorting to isolate OSCs, an approach first reported over 10 years ago20 with more than 30 corroborating studies since then (Supplementary Table S1), has retrieved PVCs. In addition, DDX4 antibodies have been used to sort PGC-like cells from cultures of human embryonic stem cells and induced pluripotent stem cells,65,66 indicating that the utility of this approach to specifically isolate primitive germ cells is not unique to OSCs. Notably, only PGC–like cells were obtained after DDX4 antibody–based sorting in these 2 studies,65,66 even though PVCs also arise in cultures of differentiating human pluripotent stem cells.67 This discordance in what Wagner et al.25 reported regarding their isolation of PVCs instead of OSCs by this approach also extends to their own previously published findings, in which identical claims were made that human OSCs do not exist and that the sorting strategy for OSC isolation using DDX4 antibodies does not work.68 While those claims were experimentally disputed,69 in this earlier study the authors similarly performed scRNA-seq on “DDX4+” cells obtained from human ovarian cortical biopsies. Their analysis in that prior study did not, however, identify PVCs as the primary cell type retrieved by FACS. Instead, out of a randomly selected population of 41 “DDX4+” cells, their gene expression associations identified a mixed population of very diverse cell types.68 The inconsistent outcomes reported by these authors when using DDX4 antibodies for cell sorting in their own studies25,68 may help explain why their findings diverge widely from what many others have consistently reported using the same cell sorting strategy since 2009 (Supplementary Table S1) (see also Supplementary material Discussion 1).
With this information as a preface, we designed experiments to determine how PVCs could be erroneously isolated as cells perceived to be antibody-positive using flow cytometry. Our data presented herein offer at least a reasonable explanation for this outcome, which accounts for the inherent autofluorescence of PVCs being detected as a false-positive signal during FACS. This would lead to an artifactual enrichment of these cells rather than true antibody-positive cells. In turn, our analysis of PVCs and non–oocyte germ cells in the Wagner et al.25 dataset demonstrated that these 2 cell types, not surprisingly, cluster separately and exhibit no overlap in gene expression profiles associated with each cell type. Thus, even if PVCs were isolated by DDX4 antibody–based FACS for reasons unrelated to endogenous autofluorescence, any downstream analysis of these cells would generate data that differ considerably from the published results from many other groups that have successfully sorted human OSCs for characterization of their germline identity and oocyte-forming properties.7-15
Conclusions
Since the initial report on OSCs almost 2 decades ago,3 over 80 primary research studies have been published supporting the existence of OSCs and/or postnatal oogenesis in mammals6 (Supplementary Tables S1–S3). More than 30 of these have sorted OSCs from ovaries with polyclonal or monoclonal antibodies directed against the C terminus of DDX4 for in-depth characterization (Supplementary Table S1). In this same time frame, only 10 primary research papers have been published disputing the existence of OSCs and/or the occurrence of postnatal oogenesis in mammals (Supplementary Tables S1 and S3), and only 4 of these studies claimed that DDX4 antibody-based sorting fails to isolate OSCs (Supplementary Table S1; see also Supplementary material Discussion 3). With respect to human OSCs, at least 6 different groups have established, and independently corroborated, that extracellular DDX4-positive cells sorted from adult human ovarian cortex express primitive germ lineage (but not oocyte) markers, can be expanded in number in culture, activate meiosis, and generate oocyte-like cells in vitro and oocytes in ovarian tissue.7-15 Aside from the fact that these outcomes are fully consistent with a large body of work on OSCs in other species,6,18,70 none of these endpoints are features of PVCs.
In consideration of this, along with the experimental evidence presented herein, a more reasonable conclusion from the Wagner et al.25 study is that the scRNA-seq workflow used by the authors was not designed appropriately to identify candidate OSCs, or in fact any stem cell type, in their sample. When the analytical workflow was optimized and applied to all cells of their sample equally, we uncovered evidence in their dataset of the existence of both OSCs and primitive germ cells entering meiosis-I. Likewise, a more reasonable conclusion from their flow cytometry work is that these authors have had recurrent technical difficulties with FACS over the years25,68 in achieving what more than 30 other studies have already reported with respect to the sorting of OSCs from adult ovarian tissue for in vitro and in vivo characterization (Supplementary Table S1). This alternative conclusion would also remove the erroneous inference made by Wagner et al.25 that numerous other groups have mistakenly worked with PVCs, and not germ cells, in the many reports of OSCs6 (Supplementary Table S1) or PGC-like cells64,65 published to date using DDX4 antibody-based sorting to isolate primitive germ lineage cells.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grant R01-AG012279 to J.L.T. The efforts of T.B. were partially supported by a Northeastern University PEAK Award.
Conflict of Interest
H.A., T.B., Y.T., O.I., H.S., R.A.A., and E.E.T.: declare no potential conflict of interests. Z.F.: declares leadership position with Foundation Medicine, Abbvie and ownership interest with Roche, Abbvie. D.C.W.: declares interest in intellectual property described in U.S. Patent 8,642,329, U.S. Patent 8,647,869, U.S. Patent 9,150,830, and U.S. Patent 10,525,086. J.L.T. declares interest in intellectual property described in U.S. Patent 7,195,775, U.S. Patent 7,850,984, U.S. Patent 7,955,846, U.S. Patent 8,642,329, U.S. Patent 8,647,869, U.S. Patent 8,652,840, U.S. Patent 9,150,830, U.S. Patent 9,267,111, U.S. Patent 9,845,482, and U.S. Patent 10,525,086.
Author Contributions
H.A., Z.F., and D.C.W: performed the analysis of the scRNA-seq dataset; H.A.: performed the flow cytometric analysis; T.B.: assisted with the experiments and performed analysis of published studies of OSCs, postnatal oogenesis, and other ovarian stem cell types; R.A.A., E.E.T. Y.T., O.I., and H.S.: collected and cryopreserved human ovarian cortical tissue for analysis; D.C.W. and J.L.T.: directed the experiments; and J.L.T.: wrote the manuscript; all authors approved the results and the final manuscript for submission.
Data Availability
The data underlying this article are available in the article and in its Supplementary material.
References
- 1. Zuckerman S. The number of oocytes in the mature ovary. Rec Prog Horm Res. 1951;6:63-108. [Google Scholar]
- 2. Fayomi AP, Orwig KE.. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207-214. https://doi.org/10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson J, Canning J, Kaneko T, et al. Germline stem cells and follicular renewal in the postnatal mammalian ovary. Nature. 2004;428(6979):145-150. https://doi.org/10.1038/nature02316. [DOI] [PubMed] [Google Scholar]
- 4. Tilly JL, Niikura Y, Rueda BR.. The current status of evidence for and against postnatal oogenesis in mammals: a case of ovarian optimism versus pessimism?. Biol Reprod. 2009;80(1):2-12. https://doi.org/10.1095/biolreprod.108.069088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woods DC, White YAR, Tilly JL.. Purification of oogonial stem cells from adult mouse and human ovaries: an assessment of the literature and a view towards the future. Reprod Sci. 2013;20(1):7-15. https://doi.org/10.1177/1933719112462632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin JJ, Woods DC, Tilly JL.. Implications and current limitations of oogenesis from female germline or oogonial stem cells in adult mammalian ovaries. Cells. 2019;8(2):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. White YA, Woods DC, Takai Y, et al. Oocyte formation by mitotically active germ cells purified from ovaries of reproductive-age women. Nat Med. 2012;18(3):413-421. https://doi.org/10.1038/nm.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Woods DC, Tilly JL.. Isolation, characterization and propagation of mitotically active germ cells from adult mouse and human ovaries. Nat Protoc. 2013;8(5):966-988. https://doi.org/10.1038/nprot.2013.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ding X, Liu G, Xu B, et al. Human GV oocytes generated by mitotically active germ cells obtained from follicular aspirates. Sci Rep. 2016;6:28218. https://doi.org/10.1038/srep28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bothun AM, Gao Y, Takai Y, et al. Quantitative proteomic profiling of the human ovary from early to mid-gestation reveals protein expression dynamics of oogenesis and folliculogenesis. Stem Cells Dev. 2018;27(11):723-735. https://doi.org/10.1089/scd.2018.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarkson YL, McLaughlin M, Waterfall M, et al. Initial characterisation of adult human ovarian cell populations isolated by DDX4 expression and aldehyde dehydrogenase activity. Sci Rep. 2018;8(1):6953. https://doi.org/10.1038/s41598-018-25116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Silvestris E, Cafforio P, D’Oronzo S, et al. In vitro differentiation of human oocyte-like cells from oogonial stem cells: single-cell isolation and molecular characterization. Hum Reprod. 2018;33(3):464-473. https://doi.org/10.1093/humrep/dex377. [DOI] [PubMed] [Google Scholar]
- 13. MacDonald JA, Takai Y, Ishihara O, et al. Extracellular matrix signaling activates differentiation of adult ovary-derived oogonial stem cells in a species-specific manner. Fertil Steril. 2019;111(4):794-805. https://doi.org/10.1016/j.fertnstert.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ariyath A, Janu MK, Paul-Prasanth B.. Differentiation potential of cultured extracellular DEAD-box helicase 4+ oogonial stem cells from adult human ovaries into somatic lineages. Cells Tissues Organs. 2021. https://doi.org/10.1159/000519087. [DOI] [PubMed] [Google Scholar]
- 15. Sequeira RC, Sittadjody S, Criswell T, et al. Enhanced method to select human oogonial stem cells for fertility research. Cell Tissue Res. 2021;386(1):145-156. https://doi.org/10.1007/s00441-021-03464-1. [DOI] [PubMed] [Google Scholar]
- 16. Woods DC, Tilly JL.. An evolutionary perspective on female germline stem cell function from flies to humans. Semin Reprod Med. 2013;31(1):24-32. https://doi.org/10.1055/s-0032-1331794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Woods DC, Tilly JL.. Autologous germline mitochondrial energy transfer (AUGMENT) in assisted human reproduction. Semin Reprod Med. 2015;33(6):410-421. https://doi.org/10.1055/s-0035-1567826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Akahori T, Woods DC, Tilly JL.. Female fertility preservation through stem cell-based ovarian tissue reconstitution in vitro and ovarian regeneration in vivo. Clin Med Insights Reprod Health. 2019;13:1179558119848007. https://doi.org/10.1177/1179558119848007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Telfer EE, Anderson RA.. The existence and potential of germline stem cells in the adult mammalian ovary. Climacteric. 2019;34(1):297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zou K, Yuan Z, Yang Z, et al. Production of offspring from a germline stem cell line derived from neonatal ovaries. Nat Cell Biol. 2009;11(5):631-636. https://doi.org/10.1038/ncb1869. [DOI] [PubMed] [Google Scholar]
- 21. Wang N, Satirapod C, Ohguchi Y, et al. Genetic studies in mice directly link oocytes produced during adulthood to ovarian function and natural fertility. Sci Rep. 2017;7(1):10011. https://doi.org/10.1038/s41598-017-10033-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fakih MH, El Shmoury M, Szeptycki J, et al. The AUGMENTSM treatment: physician reported outcomes of the initial global patient experience. JFIV Reprod Med Genet. 2015;3:154. [Google Scholar]
- 23. Oktay K, Baltaci V, Sonmezer M, et al. Oogonial precursor cell derived autologous mitochondria injection (AMI) to improve outcomes in women with multiple IVF failures due to low oocyte quality: a clinical translation. Reprod Sci. 2015;22(12):1612-1617. https://doi.org/10.1177/1933719115612137. [DOI] [PubMed] [Google Scholar]
- 24. Tilly JL, Woods DC.. The obligate need for accuracy in reporting preclinical studies relevant to clinical trials: autologous germline mitochondrial supplementation for assisted human reproduction as a case study. Ther Adv Reprod Health. 2020;14:2633494120917350. https://doi.org/10.1177/2633494120917350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagner M, Yoshihara M, Douagi I, et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11:1147. https://doi.org/10.1038/s41467-020-14936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W, Zhao Y, Chen X, et al. A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples. Nat Biotechnol. 2021;39(9):1103-1114. https://doi.org/10.1038/s41587-020-00748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pan B, Kusko R, Xiao W, et al. Similarities and differences between variants called with human reference genome HG19 or HG38. BMC Bioinf. 2019;20(Suppl 2):101. https://doi.org/10.1186/s12859-019-2620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Navaroli DM, Tilly JL, Woods DC.. Isolation of mammalian oogonial stem cells by antibody-based fluorescence-activated cell sorting. Methods Mol Biol. 2016;1457:253-268. https://doi.org/10.1007/978-1-4939-3795-0_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. MacDonald JA, Woods DC, Tilly JL.. Biomechanical strain directly activates the differentiation of murine oogonial stem cells. Stem Cells Dev. 2021;30(15):749-757. https://doi.org/10.1089/scd.2021.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Satirapod C, Wang N, MacDonald JA, et al. Estrogen regulation of female germline stem cell differentiation as a mechanism contributing to female reproductive aging. Aging (Albany, NY). 2020;12(8):7313-7333. https://doi.org/10.18632/aging.103080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo K, Li CH, Wang XY, et al. Germ stem cells are active in postnatal mouse ovary under physiological conditions. Mol Hum Reprod. 2016;22(5):316-328. https://doi.org/10.1093/molehr/gaw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma D, Bhartiya D.. Stem cells in adult mice ovaries form germ cell nests, undergo meiosis, neo-oogenesis and follicle assembly on regular basis during estrus cycle. Stem Cell Rev Rep. 2021;17(5):1695-1711. https://doi.org/10.1007/s12015-021-10237-4. [DOI] [PubMed] [Google Scholar]
- 33. de Souza GB, Costa J, da Cunha EV, et al. Bovine ovarian stem cells differentiate into germ cells and oocyte-like structures after culture in vitro. Reprod Domest Anim. 2017;52(2):243-250. https://doi.org/10.1111/rda.12886. [DOI] [PubMed] [Google Scholar]
- 34. Liang L, Soyal SM, Dean J.. FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development. 1997;124(24):4939-4947. https://doi.org/10.1242/dev.124.24.4939. [DOI] [PubMed] [Google Scholar]
- 35. Dean J. Oocyte-specific genes regulate follicle formation, fertility and early mouse development. J Reprod Immunol. 2002;53(1-2):171-180. https://doi.org/10.1016/s0165-0378(01)00100-0. [DOI] [PubMed] [Google Scholar]
- 36. Huntriss J, Gosden R, Hinkins M, et al. Isolation, characterization and expression of the human Factor In the Germline alpha (FIGLA) gene in ovarian follicles and oocytes. Mol Hum Reprod. 2002;8(12):1087-1095. https://doi.org/10.1093/molehr/8.12.1087. [DOI] [PubMed] [Google Scholar]
- 37. Wang Z, Liu C-Y, Zhao Y, et al. FIGLA, LHX8 and SOHLH1 transcription factor networks regulate mouse oocyte growth and differentiation. Nucl Acids Res. 2020;48(7):3525-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Castrillon DH, Quade BJ, Wang TY, et al. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci USA. 2000;97(17):9585-9590. https://doi.org/10.1073/pnas.160274797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Toyooka Y, Tsunekawa N, Takahashi Y, et al. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev. 2000;93(1-2):139-149. https://doi.org/10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- 40. Le HT, Hasegawa Y, Daitoku Y, et al. Generation of B6-Ddx4em1(CreERT2)Utr, a novel CreERT2 knock-in line, for germ cell lineage by CRISPR/Cas9. Genesis. 2020;58(7):e23367. https://doi.org/10.1002/dvg.23367. [DOI] [PubMed] [Google Scholar]
- 41. Suzumori N, Yan C, Matzuk MM, et al. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111(1-2):137-141. https://doi.org/10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 42. Huntriss J, Hinkins M, Picton HM.. cDNA cloning and expression of the human NOBOX gene in oocytes and ovarian follicles. Mol Hum Reprod. 2006;12(5):283-289. https://doi.org/10.1093/molehr/gal035. [DOI] [PubMed] [Google Scholar]
- 43. Heyting C. Synaptonemal complexes: structure and function. Curr Opin Cell Biol. 1996;8(3):389-396. https://doi.org/10.1016/s0955-0674(96)80015-9. [DOI] [PubMed] [Google Scholar]
- 44. Page SL, Hawley RS.. The genetics and molecular biology of the synaptonemal complex. Ann Rev Cell Dev Biol. 2004;20:525-558. [DOI] [PubMed] [Google Scholar]
- 45. Prieto I, Suja J, Pezzi N, et al. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat Cell Biol. 2001;3(8):761-766. [DOI] [PubMed] [Google Scholar]
- 46. Hopkins J, Hwang G, Jacob J, et al. Meiosis-specific cohesin component, Stag3 is essential for maintaining centromere chromatid cohesion, and required for DNA repair and synapsis between homologous chromosomes. PLoS Genet. 2014;10(7):e1004413. https://doi.org/10.1371/journal.pgen.1004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Winters T, McNicoll F, Jessberger R.. Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion. EMBO J. 2014;33(11):1256-1270. https://doi.org/10.1002/embj.201387330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishiguro KI. The cohesin complex in mammalian meiosis. Genes Cells. 2019;24(1):6-30. https://doi.org/10.1111/gtc.12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng CW, Bowles J, Koopman P.. Control of mammalian germ cell entry into meiosis. Mol Cell Endocrinol. 2014;382(1):488-497. https://doi.org/10.1016/j.mce.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 50. Yan C, Pendola FL, Jacob R, et al. Oosp1 encodes a novel mouse oocyte-secreted protein. Genesis. 2001;31(3):105-110. https://doi.org/10.1002/gene.10010. [DOI] [PubMed] [Google Scholar]
- 51. Paillisson A, Dadé S, Callebaut I, et al. Identification, characterization and metagenome analysis of oocyte-specific genes organized in clusters in the mouse genome. BMC Genomics. 2005;6:76. https://doi.org/10.1186/1471-2164-6-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397-406. https://doi.org/10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fan X, Bialecka M, Moustakas I, et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat Commun. 2019;10(1):3164. https://doi.org/10.1038/s41467-019-11036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nicholls PK, Schorle H, Naqvi S, et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc Natl Acad Sci USA. 2019;116(51):25677-25687. https://doi.org/10.1073/pnas.1910733116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yang CR, Rajkovic G, Daldello EM, et al. The RNA-binding protein DAZL functions as repressor and activator of mRNA translation during oocyte maturation. Nat Commun. 2020;11(1):1399. https://doi.org/10.1038/s41467-020-15209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol Ann Rev. 2005;11:227-256. [DOI] [PubMed] [Google Scholar]
- 57. Donnenberg VS, Donnenberg AD.. Coping with artifact in the analysis of flow cytometric data. Methods. 2015;82:3-11. https://doi.org/10.1016/j.ymeth.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 58. Choi JR, Yong KW, Choi JY, et al. Single-cell RNA sequencing and its combination with protein and DNA analyses. Cells. 2020;9(5):1130. https://doi.org/10.3390/cells9051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bhartiya D, Kausik A, Singh P, et al. Will single-cell RNAseq decipher stem cells biology in normal and cancerous tissues?. Hum Reprod Update. 2021;27(2):421. https://doi.org/10.1093/humupd/dmaa058. [DOI] [PubMed] [Google Scholar]
- 60. Saitou M, Barton SC, Surani MA.. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418(6895):293-300. [DOI] [PubMed] [Google Scholar]
- 61. Ohinata Y, Payer B, O’Carroll D, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207-213. [DOI] [PubMed] [Google Scholar]
- 62. Guo F, Yan L, Guo H, et al. The transcriptome and DNA methylome landscape of human primordial germ cells. Cell. 2015;161(6):1437-1452. [DOI] [PubMed] [Google Scholar]
- 63. Sugawa F, Araúzo-Bravo MJ, Yoon J, et al. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J. 2015;34(8):1009-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tilly JL, Woods DC.. Reproductive biology at the crossroads of stem cell biology and big-data. Fertil Steril. 2021;116(3):686-687. [DOI] [PubMed] [Google Scholar]
- 65. Richards M, Fong C-Y, Bongso A.. Comparative evaluation of different in vitro systems that stimulate germ cell differentiation in human embryonic stem cells. Fertil Steril. 2010;93(3):986-994. [DOI] [PubMed] [Google Scholar]
- 66. Yu DCW, Wu FC, Wu CE, et al. Human pluripotent stem cell derived DDX4 and KRT-8 positive cells participate in ovarian follicle-like structure formation. iScience. 2020;24(1):102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wanjare M, Kusuma S, Gerecht S.. Defining differences among perivascular cells derived from human pluripotent stem cells. Stem Cell Rep. 2014;2(5):561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang H, Panula S, Petropoulos S, et al. Adult human and mouse ovaries lack DDX4-expressing functional oogonial stem cells. Nat Med. 2015;21(10):1116-1118. [DOI] [PubMed] [Google Scholar]
- 69. Woods DC, Tilly JL.. Reply to adult human and mouse ovaries lack DDX4-expressing functional oogonial stem cells. Nat Med. 2015;21(10):1118-1121. [DOI] [PubMed] [Google Scholar]
- 70. Truman AM, Tilly JL, Woods DC.. Ovarian regeneration: the potential for stem cell contribution in the postnatal ovary to sustained endocrine function. Mol Cell Endocrinol. 2017;445:74-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data referenced in this study were originally generated by Wagner et al.25 from adult human ovarian cortical biopsies of 4 subjects (CSP, n = 3; GRP, n = 1). The 10× Genomics dataset of Wagner et al.25 for adult human unsorted ovarian cortical cells was deposited by these authors to, and accessed by us through, the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) under the accession code E-MTAb−8381. Analyses of scRNA-seq data were completed using the lines of code for adult human unsorted ovarian cortical cells deposited by Wagner et al.25 on GitHub (https://github.com/wagmag/SingleCellOvary). For Cell Ranger v6 analysis, additional lines of code were run in parallel to select proper quality control metrics as well as to determine the parameters for dimensionality reduction that best represented the data. The code used with the Cell Ranger v6 analysis is available on GitHub (https://github.com/hanrico/Ovarian-scRNA-seq).
The data underlying this article are available in the article and in its Supplementary material.