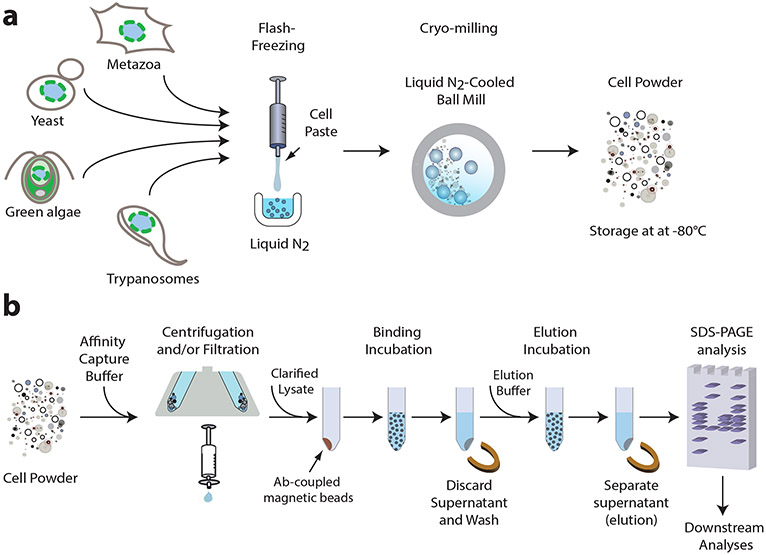
Fig. 1. Affinity isolation methodology.
An outline of a general affinity isolation protocol, which can be optimized for various protein complexes from many organisms, as described in this work. (a) Cell lysis using flash freezing and cryo-milling to obtain cell powder which can be stored at −80°C and used as needed for any scale experiment. (b) Affinity capture by resuspending cell powder in an optimized affinity capture buffer and using the clarified (centrifuged and/or filtered) lysate for incubation with antibody-coupled magnetic beads to capture the complex on the beads followed by discarding the remaining supernatant and washing steps. Complex elution is accomplished by denaturation or by native elution (proteolytic removal or competitive binding) followed by separation from the beads. Eluted complex can be analyzed by SDS-PAGE and other downstream analyses mentioned in the text.