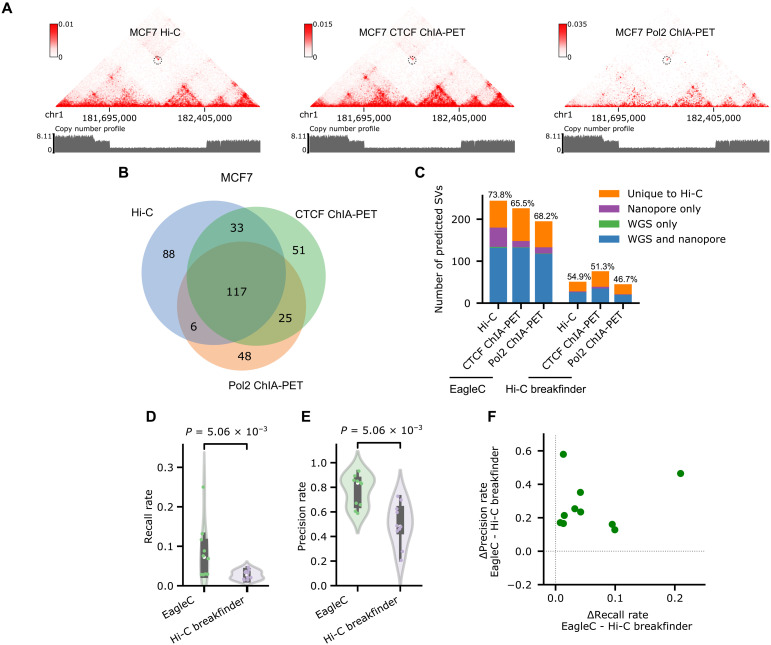
Fig. 4. EagleC accurately predicts SVs on HiChIP and ChIA-PET contact maps.
(A) A heterozygous deletion (chr1, 181,695,000 to 182,405,000) on chromosome 1 is correctly predicted by EagleC on Hi-C, CTCF ChIA-PET, and Pol2 ChIA-PET contact maps of MCF7 cells. The same copy number profile calculated from WGS is shown below each contact map. (B) Venn diagram of SVs predicted by EagleC on different contact maps of MCF7 cells. (C) Number of SVs predicted by Hi-C breakfinder and EagleC on different contact maps of MCF7 cells with levels of validation by orthogonal methods. The ratio of SVs that can be validated by orthogonal methods is indicated above each bar. (D and E) Performance of EagleC and Hi-C breakfinder on 10 HiChIP/ChIA-PET contact maps in cancer samples. Each dot represents an individual sample. The P values were computed using the two-sided Wilcoxon signed-rank test. (F) Differences of precision rates and recall rates between EagleC-predicted and Hi-C breakfinder–predicted SVs on 10 HiChIP/ChIA-PET datasets in cancer.