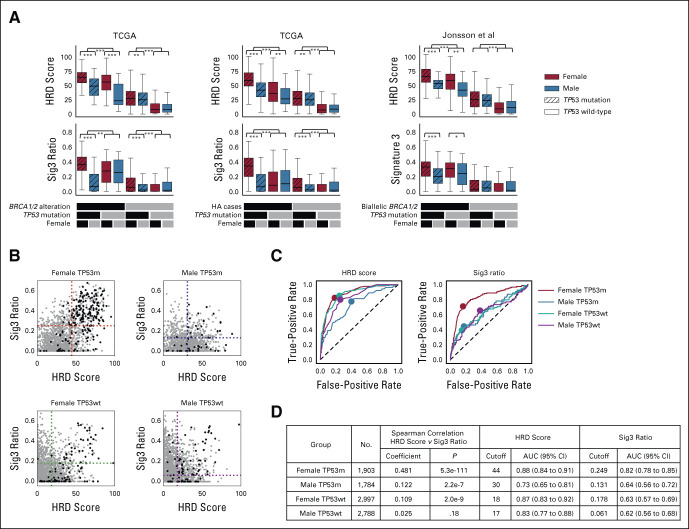
FIG 3.
Systematization and results for GS score–based classification. (A) Comparison of HRD score and Sig3 ratio stratified by TP53 mutation and sex. Further stratification is applied using biallelic BRCA1/2 alteration (including BRCA1 methylation, left), biallelic HRR pathway gene alteration (middle), biallelic BRCA1/2 alteration in Jonsson's data set (right). *, **, and *** stand for P < .05, P < .01, and P < .001 in the Mann-Whitney U test, respectively. (B) HRD score and Sig3 ratio pairwise distribution when patients are divided by sex and TP53 mutation status. Black dots represent HRR pathway gene-altered cases, and gray dots indicate nonaltered cases. Dotted lines are placed at the optimal cutoff values determined by ROC curve analysis. (C) Differences in ROC curves and optimal cutoff values for HRR pathway gene alterations in HRD score and Sig3 ratio among the four groups. (D) Per stratification, Spearman correlation coefficients between the HRD score and the Sig3 ratio, optimized scar score cutoff values, and resulting AUCs are shown. AUC, area under the curve; GS, genomic scar high; HRD, homologous recombination deficiency; HRR, homologous recombination repair; ROC, receiver operating characteristic; Sig3, mutational signature 3; TCGA, The Cancer Genome Atlas.