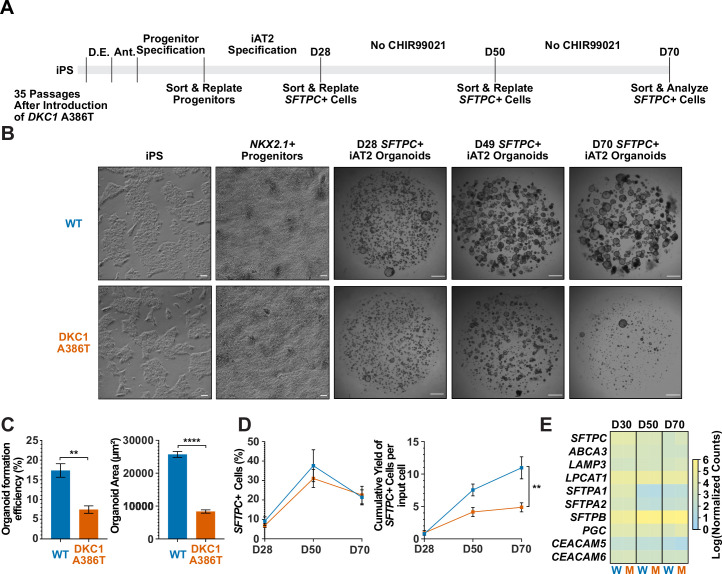
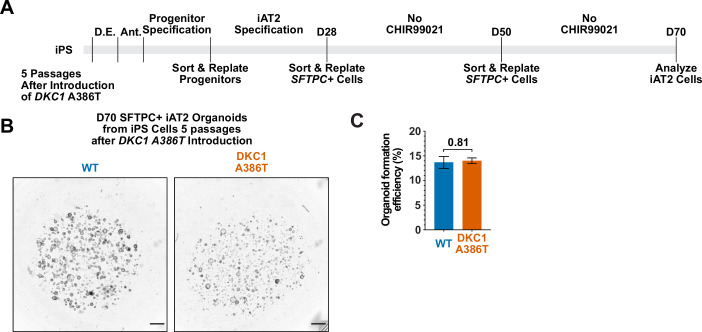
Figure 1. Dyskeratosis congenita induced pluripotent stem cell (iPSC)-derived type II alveolar epithelial cells (iAT2s) fail to form alveolospheres with successive passaging.
(A) Differentiation protocol used to probe the effects of the DKC1 A386T mutation on iAT2 cells. D.E., definitive endoderm specification; Ant., anteriorization. (B) Representative images of differentiating wild type and DKC1 A386T mutant bearing iAT2 alveolospheres (scale bars, 100 µm as indicated for iPS and NKX2.1+ progenitors; 1 mm for all alveolosphere images). (C) Quantifications of alveolosphere area and formation efficiency on D70 (n = 4; ** p<0.01, **** p<0.0001, and Student’s t-test). (D) Quantification of the percentage SFTPC+ cells and the number of SFTPC+ cells produced with passage of the iAT2 cells shows DKC1 A386T iAT2 alveolospheres accumulate fewer SFTPC+ cells (n = 4; ** p<0.01, and Student’s t-test). (E) RNA-seq of sorted SFTPC+ iAT2 cells at different passages shows AT2 cell genes are not grossly affected by the DKC1 A386T mutation (n = 4, W = wild type, M = mutant).