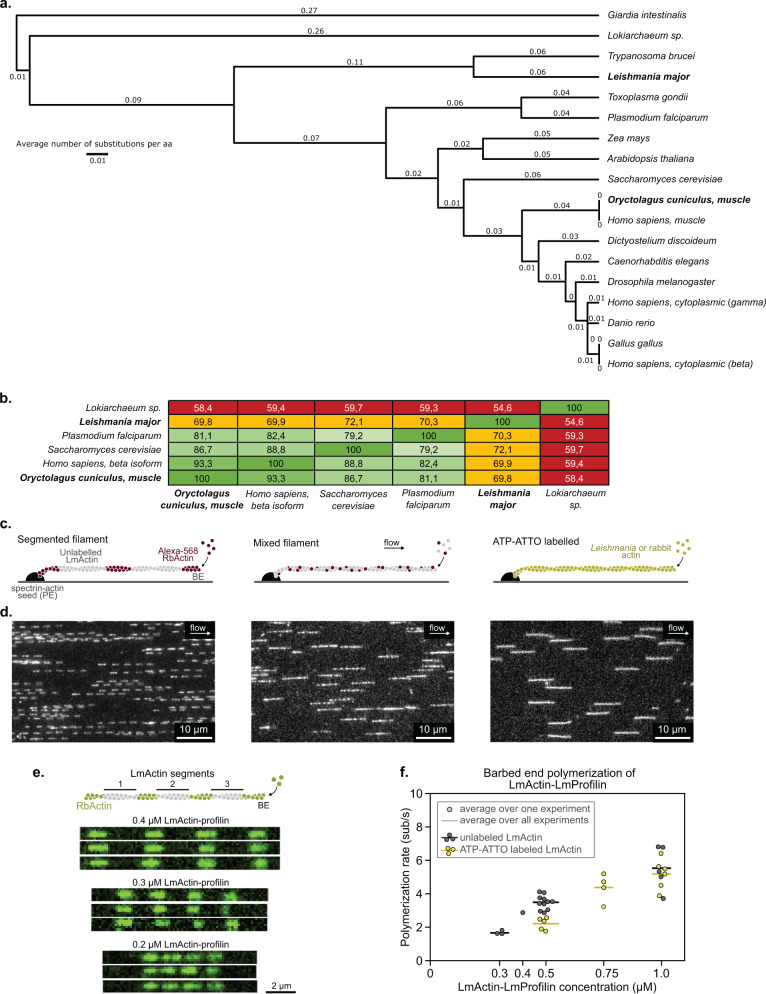
Fig. 1. Polymerization kinetics of the divergent Leishmania major actin.
a Phylogenetic tree of selected evolutionarily divergent actins based on amino acid sequences. Leishmania major parasite actin (LmActin) and α-skeletal muscle rabbit actin (Oryctolagus cuniculus, RbActin) used in this study are highlighted in bold. See the Source Data file for the sequence accession codes from Uniprot. b Pairwise comparison of the sequence identity between selected actins. c Methods applied to assemble and visualize LmActin filaments in microfluidics-coupled TIRF microscopy experiments. BE barbed end, PE pointed end. d Examples of observed actin filaments assembled using the methods described in panel c. e Typical “segmented” filaments assembled by alternating between a solution of Alexa-labeled RbActin (green) and a solution containing equimolar concentrations of unlabeled Lm-G-actin and LmProfilin. For each filament, the three LmActin segments were polymerized for 5 min with the same solution containing either 0.2, 0.3, or 0.4 µM LmActin-LmProfilin. With 0.2 µM LmActin-LmProfilin, the resulting segments were not long enough to measure the polymerization rate accurately. f Quantification of the barbed end polymerization rates with equimolar LmActin and LmProfilin, either unlabeled (in segmented filaments, gray) or labeled with 1–4 µM ATP-ATTO-488 (yellow). The polymerization rate increases with LmActin:LmProfilin concentration. Please note that especially at higher profilin:actin concentrations there is some divergence in the polymerization rates between individual experiments, and this is most likely due to spontaneous nucleation of LmActin in solution. Each data point represents an average over all measured filaments (n) from independent experiments (N), and lines correspond to the average over all experiments. For unlabeled LmActin: 0.3 µM N = 3, n = 60, 60, 60; 0.4 µM N = 1, n = 60; 0.5 µM N = 11, n = 50, 40, 40, 50, 60, 20, 20, 20, 20, 20, 10; 1 µM N = 5, n = 20, 20, 30, 40, 40. For ATP-ATTO labeled LmActin: 0.5 µM N = 5, n = 20, 20, 20, 20, 30; 0.75 µM N = 4, n = 20, 20, 20, 20; 1 µM N = 6, n = 30, 30, 20, 20, 16, 20.