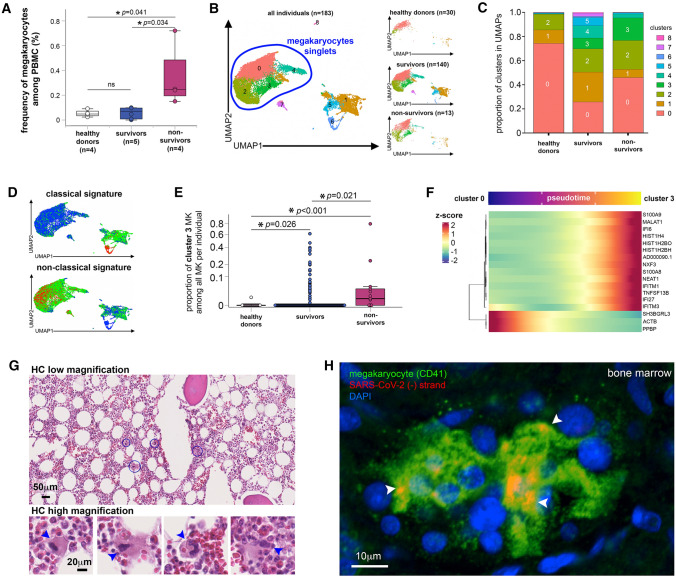
Fig. 2.
In COVID-19 non-survivors, MKs are infected and express viral sensing genes. A Frequency of MKs detected among PBMC from healthy donors and COVID-19 survivors and non-survivors as quantified by flow cytometry. Asterisks indicate statistical significance (Kruskal–Wallis test). B–F Transcriptional identity of MKs in COVID-19 patients by single-cell RNA sequencing reveals distinct phenotypes in non-survivor patients. B UMAP of single-cell transcriptomic data of MKs detected among PBMC from non-COVID-19 healthy donors (n = 30), COVID-19 survivors (n = 140) and COVID-19 non-survivors (n = 13). Unsupervised clustering detected 9 different clusters (0 to 8) of all cells analyzed. MK singlets are indicated by blue region. C Proportion of each cluster in healthy donors, COVID-19 survivors and non-survivors. D Scored gene signature expression of classical (upper) and non-classical (bottom) MK differentiation. E Fraction of cells from cluster 3 in comparison to all other clusters in individual patient samples categorized as healthy donors, COVID-19 survivors and non-survivors. F Heatmap of the genes that significantly change along pseudotime trajectory of MK development (p < 0.05 and Morans I score > 0.25). G Hematoxylin/eosin histology of bone marrow tissue obtained after COVID-19 non-survivor autopsy (low magnification (bar = 50 μm)) in which some MKs surrounded by blue circles are shown in high magnification insets (bar = 20 μm). Arrowheads indicate MKs. H Representative confocal microscopy images after CD41 (green) immunolabeling and replicative SARS-CoV-2 (-) RNA strand in situ hybridization (red) in bone marrow samples obtained from tissue autopsies of three different COVID-19 non-survivors (bar = 10 μm). Arrowheads indicate SARS-CoV-2 (-) RNA inside MKs