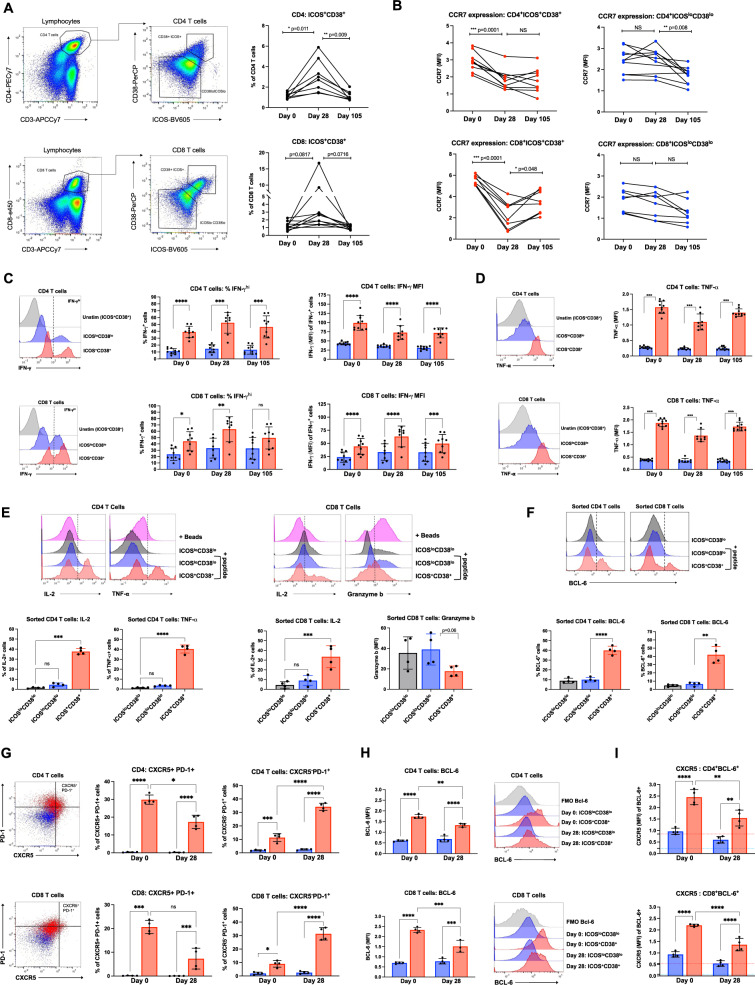
Fig. 3. Functional analysis of ICOS+CD38+ CD4+ and CD8+ T cells and response to SARS-CoV-2 Spike antigen.
PBMCs collected from study participants (n = 10) pre-vaccination (day 0), day 28 post-vaccination (7 days post-boost), and day 105 post-vaccination were examined for ICOS, CD38, and other markers by fluorescence flow cytometry individually in one setting to avoid batch variation. A CD4+ T cells were gated from lymphocytes and defined as ICOS+CD38+ or ICOSloCD38lo. Percentages of CD4+ICOS+CD38+ T cells were quantified and data from each participant is connected by lines. B Mean fluorescence intensity (MFI) of CCR7 within CD4+ICOS+CD38+, CD8+ICOS+CD38+, CD4+ICOSloCD38lo, or CD8+ICOSloCD38lo T cells. C, D PBMCs were stimulated with PMA/ionomycin and examined for cytokine production. T cells defined as ICOS+CD38+ (red) or ICOSloCD38lo (blue) were examined for C IFN-γ or D TNF. Representative samples on the left and individual participants are shown on right. E Day 28 PBMC samples were stimulated with activation beads or T cells were sorted based on CD38 and ICOS expression. Sorted T cells were labeled with CellTrace Violet (CTV) to track and stimulated with T-cell (CD3)-depleted autologous PBMCs ± SARS-CoV-2 Spike peptide pool (+ peptide, blue and red). Sorted T-cell populations were analyzed after 48 h for IL-2, TNF, and Granzyme b in F. BCL6 expression in sorted T-cell populations, stimulated as in E. G Tfh markers CXCR5 and PD-1 compared on day 0 and 28 samples. H Unstimulated days 0 and 28 ICOS+CD38+ and ICOSloCD38lo T cells were analyzed for BCL6 expression (FMO; fluorescence-minus-one control for BCL6). I CXCR5 MFI within BCL6+ T cells. FMO control for BCL6 in ICOSloCD38lo T cells is shown by the blue dotted line, and by the red dotted line for ICOS+CD38+ T cells. B Significance was determined by paired two-way ANOVA mixed-effects model. E, F Repeated measures one-way ANOVA. C, D, G–I Two-way ANOVA with Sidak’s multiple comparisons test. Representative samples are shown and each data point represents an individual participant. All error bars ± SD. ns = not significant, *p < 0.5, **p < 0.005, ***p < 0.0005, ****p < 0.00005.