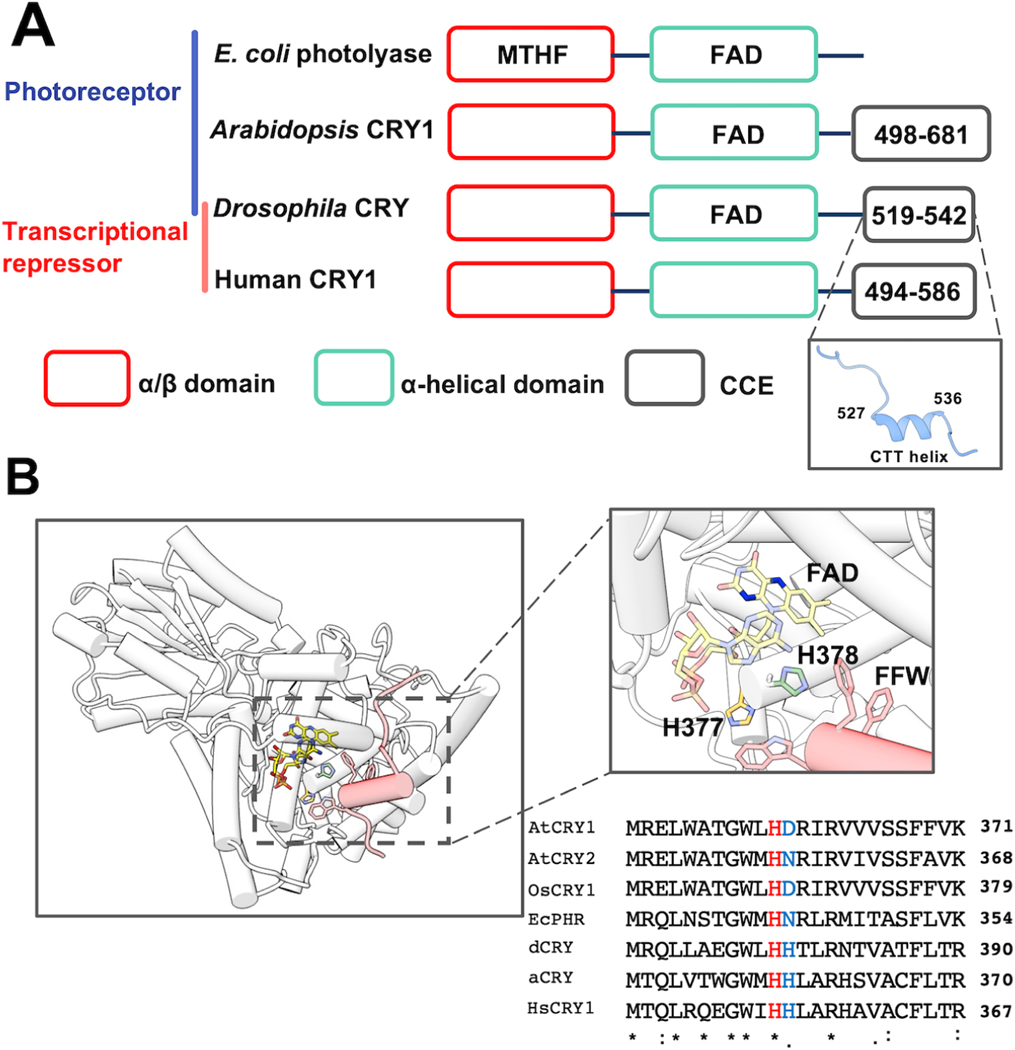
Figure 1: The flavin-pocket and CTT of Drosophila cryptochrome (dCRY).
(A) Region maps of cryptochromes and E. coli photolyases. The proteins share a photolyase homology region (PHR) comprising a conserved α/β domain (red) that binds the antenna molecule MTHF in photolyases and α-helical domain (green) that contains the FAD-binding pocket. Cryptochromes have a Cryptochrome C-terminal Extension (CCE, gray) of variable lengths. In dCRY, the CCE forms a CTT helix structure. E. coli photolyase and Type I cryptochrome Arabidopsis CRY1 are photoreceptive, whereas Type II cryptochromes such as human CRY1 function as transcriptional repressors in the circadian clock; dCRY may have both roles in flies. (B) Residues His377 (orange, with nitrogen colored blue) and His378 (green, with nitrogen colored blue) are located between cofactor FAD (colored by heteroatoms, nitrogen, light blue; oxygen, red; phosphorus, orange; N1 and N5 of the isoalloxazine ring which are involved in reduction reactions, dark blue) and FFW motif (Phe534, Phe535, Trp536, red) of the CCE (red, helices shown as cylinders) in CRY (PDB: 4GU5). The amino acid sequence of CRY in this region is aligned with other cryptochrome/photolyase proteins, such as Arabidopsis thaliana CRY1/2 (AtCRY1/2), E. coli photolyase (EcPHR), Homo sapiens CRY1 (HsCRY1), Chlamydomonas reinhardtii animal-like cryptochrome (aCRY), and Oryuza sativa japonica CRY1 (OsCRY1).