Fig. 2. Methylation profiling, copy number variations, and differentially methylated genes of low-grade and high-grade areas in DIA/DIGs.
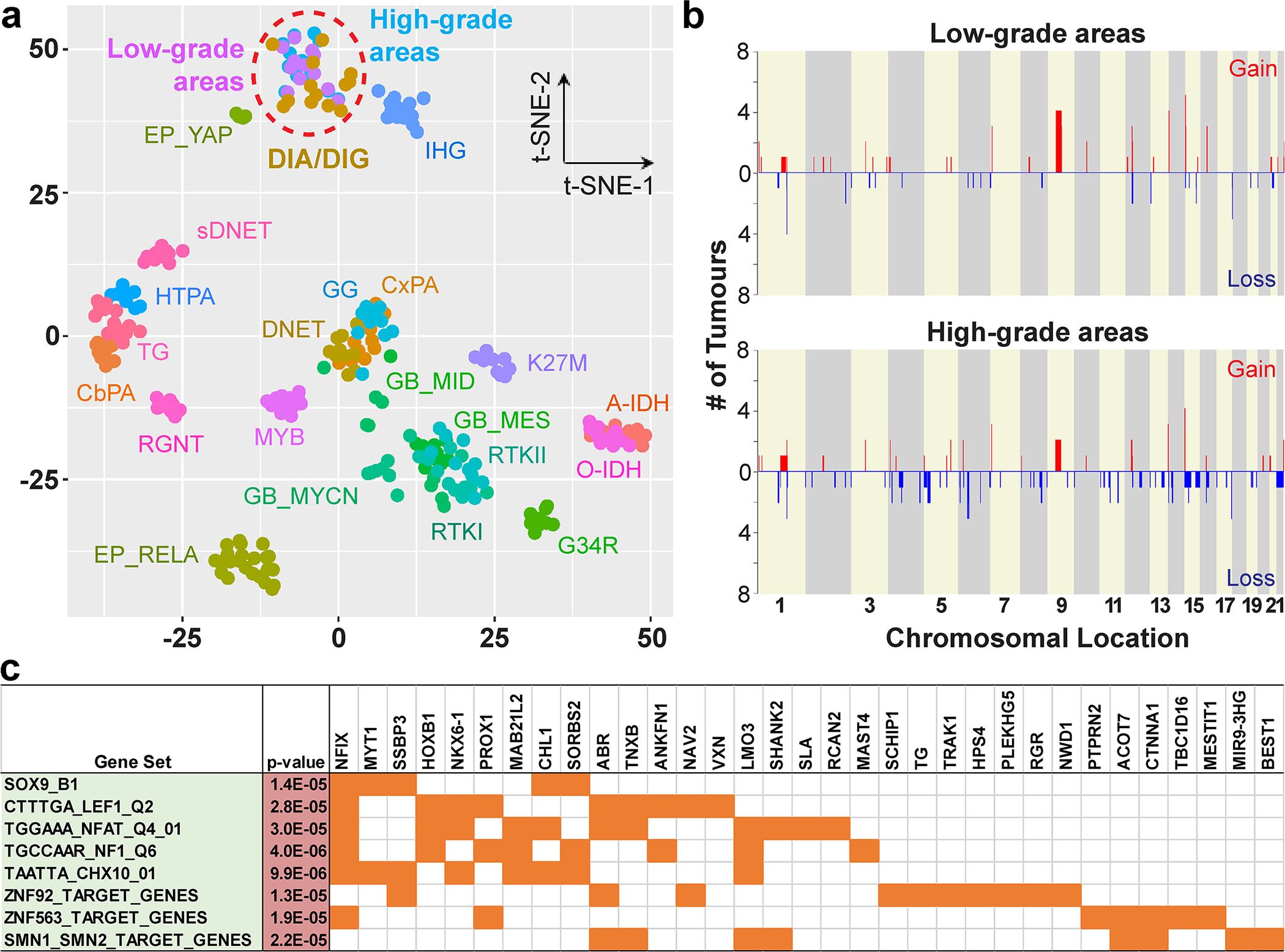
(a) A t-SNE plot shows the methylation profiles of low-grade and high-grade areas in our series of DIA/DIGs and those of reference tumours. A-IDH: IDH1-mutant diffuse astrocytoma; CbPA: cerebellar pilocytic astrocytoma; CxPA: cerebral hemispheric pilocytic astrocytoma; DNET: dysembryoplastic neuroepithelial tumour; EP: ependymoma; GB: glioblastoma; G34R: H3F3A G34R-mutant diffuse glioma; GG: ganglioglioma; HTPA: hypothalamic pilocytic astrocytoma; K27M: H3 K27M-mutant diffuse midline glioma; MYB: MYB-altered diffuse glioma; O-IDH: IDH1-mutant and 1p/19q-codeleted oligodendroglioma; RGNT: rosette-forming glioneuronal tumour; RTK I, RTK II, MES, MID, and MYCN: specific molecular groups of glioblastoma; sDNET: septal dysembryoplastic neuroepithelial tumour / myxoid glioneuronal tumour; TG: tectal glioma. (b) CNV plots of low-grade and high-grade areas in DIA/DIGs. X-axis, chromosomal location; Y-axis, number of tumours containing the gain or loss. (c) Differentially hypomethylated regions in the high-grade areas are enriched for genes downstream of specific transcription factors, including SOX9 and LEF1.
