Fig. 5. Phylogenetic associations between intact proviruses from ECs and ART-treated individuals.
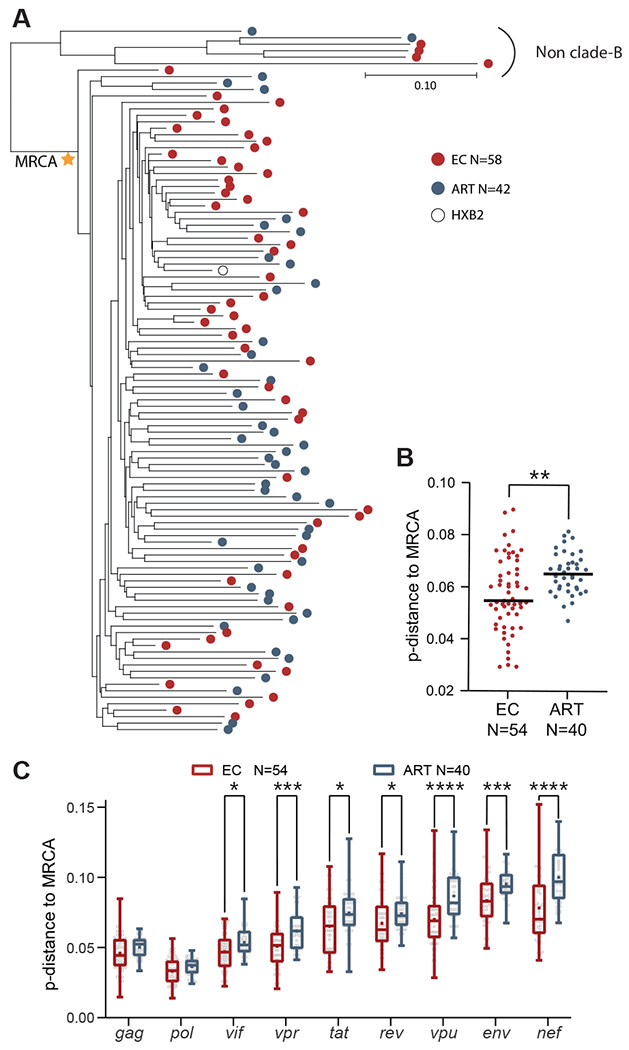
(A) Maximum likelihood phylogenetic trees of intact proviruses from ECs and ART-treated individuals are shown. The most recent common ancestor (MRCA) among clade B intact sequences is marked by a star. HXB2, reference HIV-1 clade-B sequence. (B and C) Genetic distance of all intact clade B sequences to the MRCA is shown. The MRCA and maximum likelihood-corrected distances to the MRCA were estimated in DIVEIN (71). (B) shows data for all intact proviral sequences; (C) shows data for individual HIV-1 genes within intact proviruses. An intra-individual consensus sequence reflecting the dominant base pair residue from all available intact sequences at each base pair position for each study participants was entered into the analysis in (A to C); each dot reflects one study participant. Dot plots with median are shown. Box-and-whisker plots reflect mean, median, minimum, maximum, and interquartile ranges. A two-tailed Mann Whitney U test was used in (B). An FDR-adjusted two-tailed Mann Whitney U test was used in (C). * P < 0.05, ** P < 0.01, *** P < 0.001, ****P < 0.0001.
