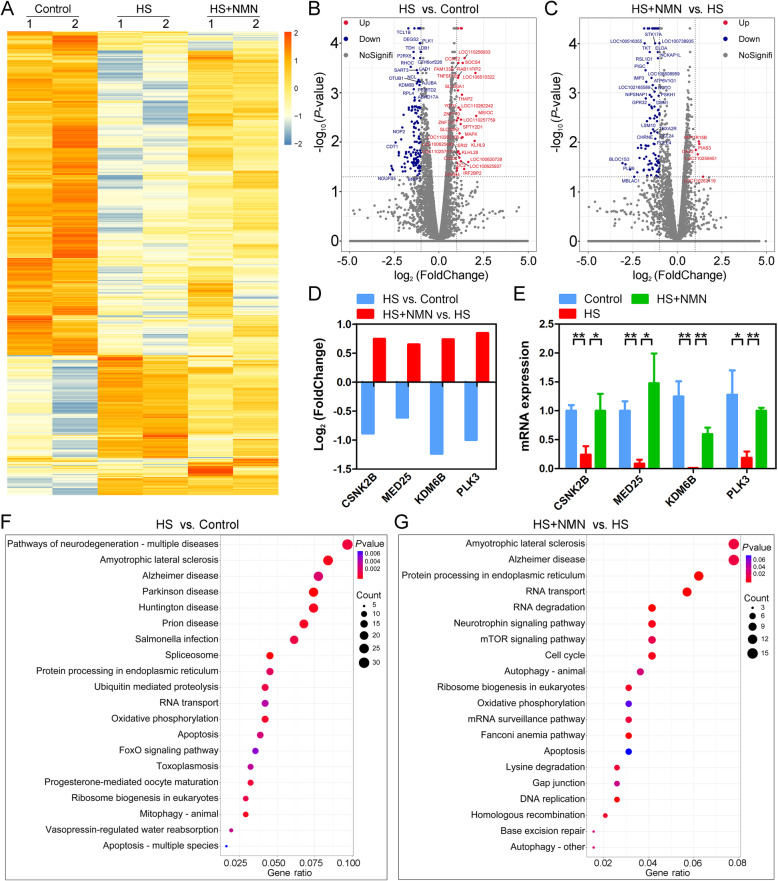
Fig. 6.
Effects of NMN supplementation on the transcriptome profiling of heat-stressed porcine oocytes. A Heatmap illustration showed the differentially expressed genes (DEGs) in control, heat-stressed and NMN-supplemented oocytes. B Volcano plot displayed the DEGs in heat-stressed oocytes compared to control oocytes. Red, upregulated; Blue, downregulated; Grey, not significant. C Volcano plot displayed the DEGs in NMN-supplemented oocytes compared to heat-stressed oocytes. Red, upregulated; Blue, downregulated; Grey, not significant. D RNA-seq results of selected genes in heat-stressed oocytes compared to control oocytes and NMN-supplemented oocytes compared to heat-stressed oocytes. E Validation of RNA-seq data by quantitative RT-PCR in control (blue), heat-stressed (red) and NMN-supplemented (green) oocytes. Data were presented as mean value (mean ± SEM) of at least three independent experiments. *P < 0.05, **P < 0.01. F KEGG pathway enrichment analysis of DEGs in heat-stressed oocytes compared to control oocytes. G KEGG pathway enrichment analysis of DEGs in NMN-supplemented oocytes compared to heat-stressed oocytes