Figure. snRNA-seq analysis identifies six cardiomyocyte subpopulations in injured and uninjured pig hearts.
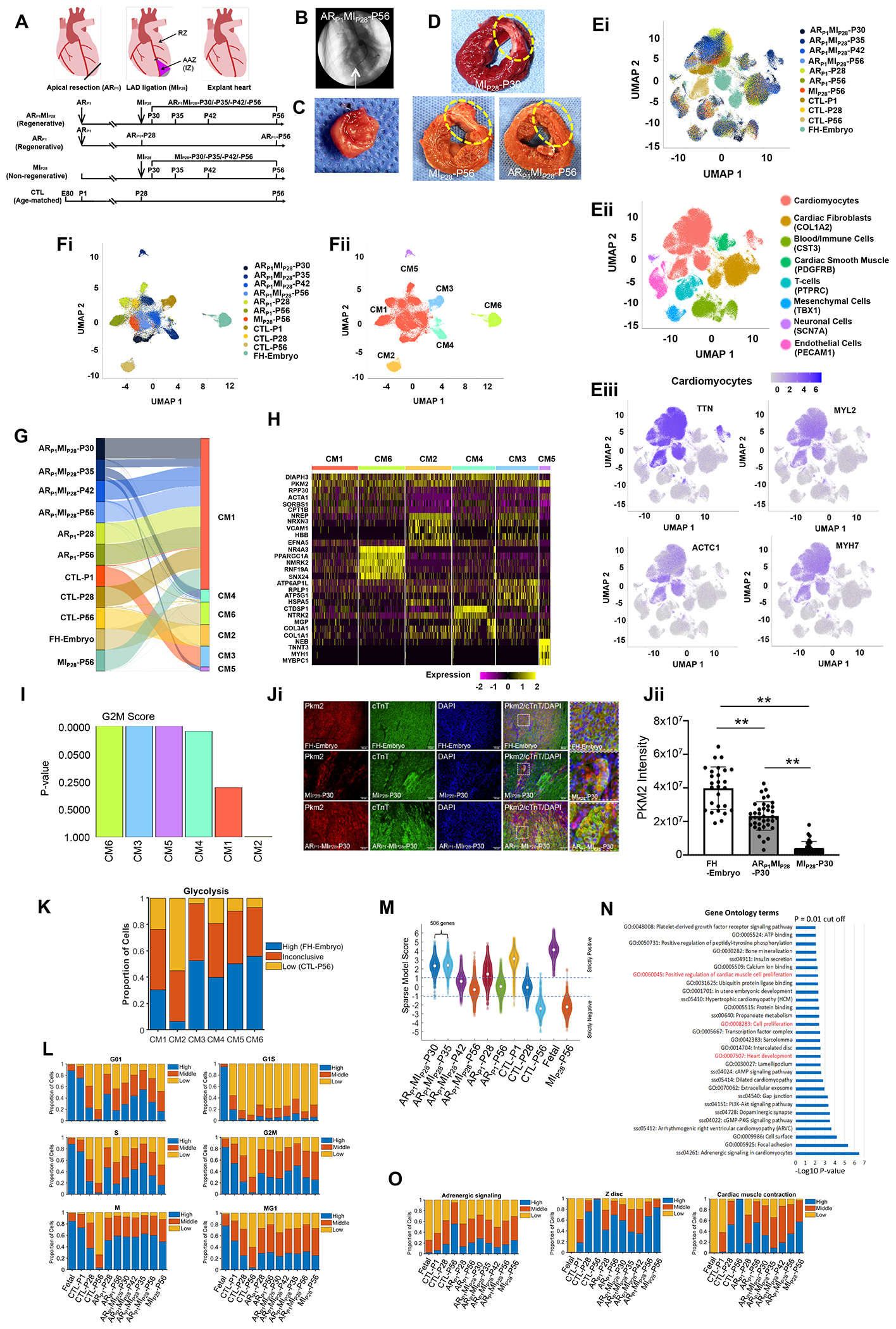
(A) Timeline of experimental procedures. (B) Coronary angiographic image with identification of the ligation site (arrow). (C-D) Images of (C) the resected apex and (D) left-ventricular sections; infarcted region is circled. (E-F) Cluster analysis for snRNA-seq data from (E) all cells and (F) cardiomyocytes; results are presented according to (Ei, Fi) experimental group/time-point, (Eii) cell type (defined by marker expression), (Eiii) abundance of each of four cardiomyocyte-specific genes, and (Fii) cardiomyocyte cluster. (G) Distribution of cardiomyocytes from each experimental group/time-point across CM clusters. (H) Heat map of expression levels for genes that were explicitly associated with each CM cluster. (I) The expression of genes associated with each cell cycle phase was analyzed and used to calculate a G2M-phase probability score for each CM cluster. (Ji) Representative images from each section FH-Embryo, ARP1MIP28-P30, and MIP28-P30 were stained with PKM2 antibody, cardiac Troponin T, and DAPI. (Jii) Quantification of PKM2 intensity by Image J. n=3 each. Square dots are for high magnification. The scale bar is 50 um. **P<0.01. (K) The expression of glycolysis genes in each CM cluster was compared to their expression in high (FH-embryo) and low (CTL-P56) cardiomyocytes. (L) The proportion of cardiomyocytes with high, medium, and low levels of expression for genes associated with each phase of the cell cycle. (M) Violin plot of the sparse model score. (N) Pathways that were significantly upregulated in ARP1MIP28-P30 and -P35 cardiomyocytes were identified via Gene Ontology Analysis. (O) The proportion of cardiomyocytes with high, medium, and low levels of expression for genes associated with adrenergic signaling, Z-disc development, and contractile activity.
