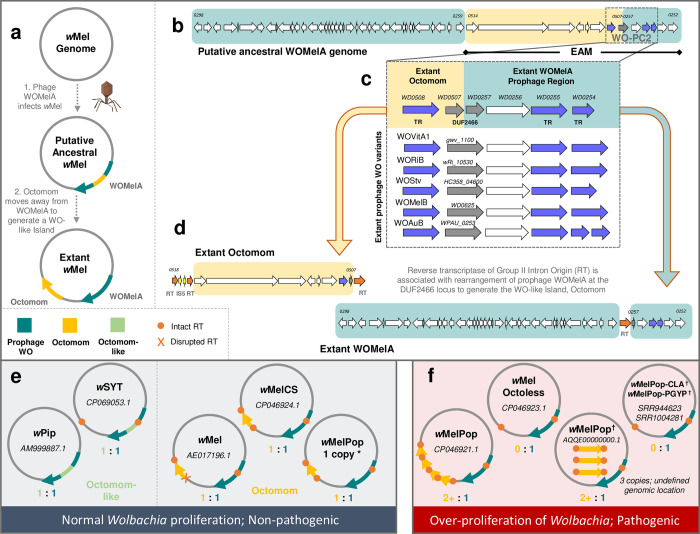
Fig 4. Comparative genomics supports a WO:Octomom origin model for Wolbachia proliferation in wMelPop.
(a) A new model for Octomom origin predicts the initial infection of wMel with a WOMelA phage. After integration, Octomom splits from the WOMelA core prophage region to form a WO-like Island. (b) A genome map of the putative, intact, ancestral WOMelA where Octomom is highlighted in yellow and the extant WOMelA genome in teal illustrates placement of Octomom in the WO EAM. (c-d) An alignment of the WO-PC2 region with closely related prophages shows that half of the conserved module (WD0507-WD0508) is now associated with Octomom and the other half (WD0257-WD0254) remained with WOMelA prophage region. DUF2466 is split across the genomic regions and, when concatenated, shares homology to intact DUF2466 genes of WO-PC2 (see S11 Fig). An IS5 insertion (d) is associated with single-copy Octomom stability in the wMel chromosome. In wMelCS-like genomes, where the flanking RTs are intact (see S10 Fig), Octomom varies in copy number. (e) When Octomom (orange-yellow) and Octomom-like (green, defined by homology to WD0512, WD0513 and WO-PC2; illustrated in S10 Fig) regions exist in a single copy, either within or outside the corresponding prophage region, Wolbachia proliferation is normal, and it is non-pathogenic. (f) If the WO-like Island occurs in multiple copies or is absent from the genome, Wolbachia over-proliferate and are pathogenic. (*) Restoring the 1:1 (WO:Octomom) ratio returns the wMelPop phenotype back to normal levels. The association of Octomom with pathogenicity (i.e., correlation vs. causation) is still to be determined [66–68]. NCBI accession numbers are listed for each genome; (†) indicates circular genomes are unavailable and genomic locations are putative.