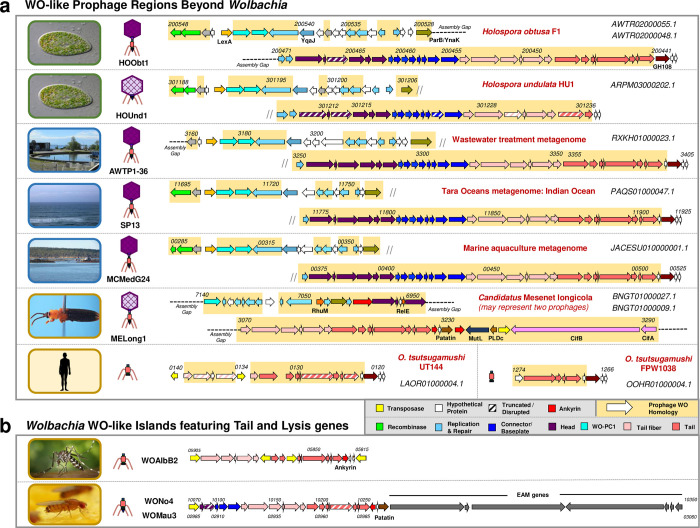
Fig 6. WO-like prophage regions are found in endonuclear Paramecium endosymbionts, aquatic environments, and other animal-associated bacteria.
(a) Genome maps of non-Wolbachia prophage regions illustrate similar gene content and synteny to prophage WO. Locus tags are listed in italics above the genes; NCBI contig accession numbers are shown in the right-hand corner of each genome. Dashed lines represent breaks in the assembly whereas small diagonal lines represent a continuation of the genome onto the next line. Genes with nucleotide homology to prophage WO are highlighted in yellow and genes of similar function are similarly color-coded according to the figure legend. Candidatus Mesenet longicola is the only genome to feature EAM genes, including cifA and cifB. Arrows with diagonal stripes represent genes that may be pseudogenized relative to homologs in other prophage genomes. Genome maps for H. elegans and H. curviuscula prophages are not shown. (b) WO-like Islands featuring tail and lysis genes share homology with the Orientia regions and may represent phage-derived bacteriocins. Predicted physical structures are illustrated to the left of each genome. Images illustrate the isolation source for each prophage: green borders represent protozoa; blue borders represent aquatic environments; gold borders represent animals. All images are available under creative commons or public domain; attribution information is provided in S8 Table.