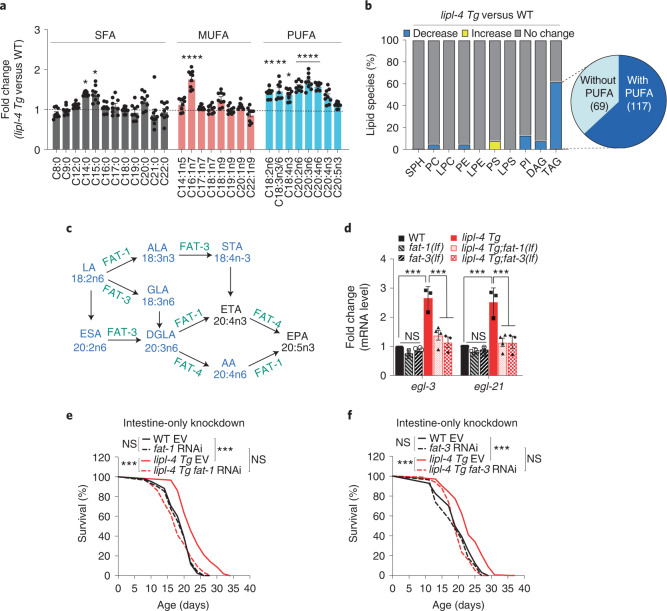
Fig. 3. Lysosome-derived PUFAs in the periphery regulate neuropeptide and longevity.
a, Relative levels of FFAs, saturated fatty acids (SFAs), MUFAs and PUFAs are quantified by liquid chromatography–mass spectrometry in lipl-4 Tg versus WT. b, Percentage of lipid species showing significant changes (P < 0.05) in the lysosome purified from lipl-4 Tg versus WT worms. Out of 186 TAG species decreased in lipl-4 Tg, 117 contain PUFAs. Sphingophospholipids (SPH), phosphatidylcholines (PC), lysophosphatidylcholines (LPC), phosphatidylethanolamines (PE), lysophosphatidylethanolamines (LPE), phosphatidylserines (PS), lysophosphatidylserines (LPS), phosphatidylinositols (PI), diacylglycerols (DAG) and TAG. c, Schematic diagram of PUFA biosynthesis in C. elegans. PUFAs enriched in lipl-4 Tg are highlighted in blue, while desaturase enzymes are marked in green. d, The loss-of-function mutation of the fatty acid desaturase fat-1(lf) or fat-3(lf) suppresses the transcriptional upregulation of egl-3 and egl-21 neuropeptide genes by lipl-4 Tg. e,f, RNAi knockdown of either fat-1 (e) or fat-3 (f) selectively in the intestine suppresses lipl-4 Tg longevity. In a, error bars represent mean ± s.e.m., n = 6 (WT) and n = 9 (lipl-4 Tg) biologically independent samples, *P = 0.028 for C18:4n3, P = 0.023 for C14:0 and P = 0.026 for C15:0, **P = 0.007 for C18:2n6 and P = 0.002 for C18:3n3/6, ****P < 0.0001 for C16:1n7, C20:2n6, C20:3n6 and C20:4n6 by two-way ANOVA with Holm–Sidak correction (lipl-4 Tg versus WT), ~40,000 worms per replicate. In d, error bars represent mean ± s.e.m., n = 4 (WT, fat-1(lf), fat-3(lf) and lipl-4 Tg;fat-1(lf)) and n = 3 (lipl-4 Tg and lipl-4 Tg;fat-3(lf)) biologically independent samples, ***P < 0.001 by two-way ANOVA with Holm–Sidak correction, ~3,000 worms per replicate. In e and f, n = 3 biologically independent samples, ***P < 0.001 by log-rank test, 60–120 worms per replicate. See Supplementary Table 5 for full lifespan data. Source numerical data are available in source data.