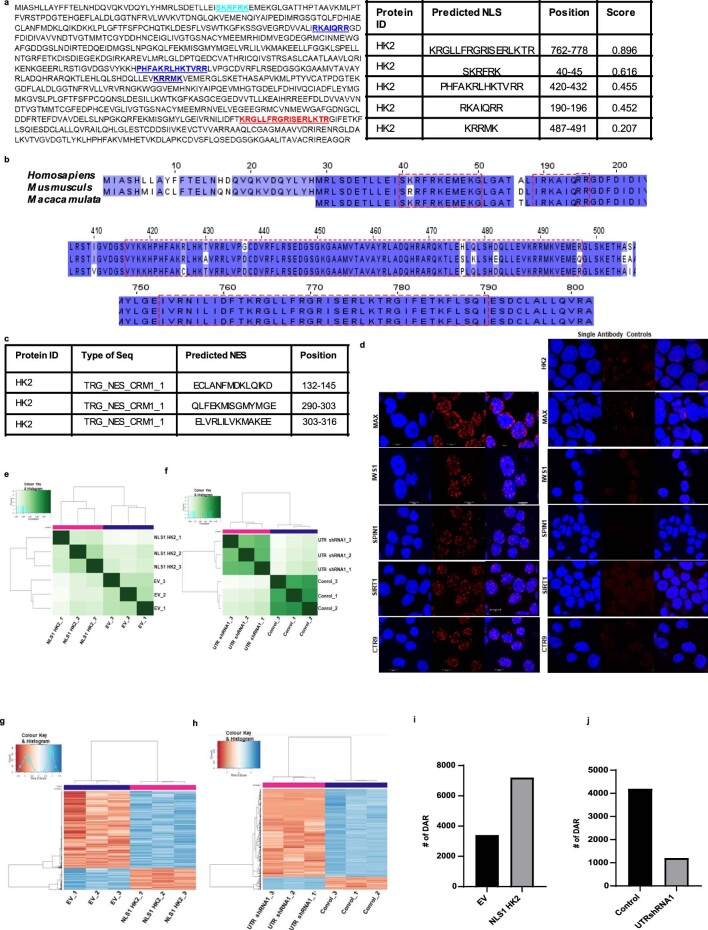
Extended Data Fig. 6. Nuclear HK2 modifies chromatin accessibility.
(a)The mammalian HK2 protein sequence was evaluated for nuclear localizing signals using the SeqNLS algorithm. (b) NLS conservation in HK2 of Homosapiens, Mus Musculus and Macaca mulata analysed using Clustal omega. (c)The predicted sequences and the positions of HK2 Nuclear Export Sequence by NESX1 library using Pattinprot and ELM library. (d) Representative confocal images of DuoLink Promity Assay interactions. Scale bar = 10µm. Representative image from 3 biologic repeats. (e) Clustering on peaks using DiffBind comparing NB4 cells transduced with NLS1-HK2 and EV. (f) Clustering on peaks using DiffBind comparing NB4 cells overexpressing OMMLS HK2 transduced with control sequences or shRNA targeting the UTR of HK2. (g) Heatmap plots of differentially accessible regions in NB4 cells overexpressing NLS1-HK2 or EV. (h) Heatmap plots of differentially accessible regions in NB4 cells overexpressing OMMLS HK2 transduced with control sequences or shRNA targeting the UTR of HK2. (i) Number of Differentially Accessible Regions in NB4 cells with NLS1-HK2 or EV measured by ATAC sequencing. (j) Number of Differentially Accessible Regions in NB4 cells overexpressing OMMLS HK2 transduced with control sequences or shRNA targeting the UTR of HK2 measured by ATAC sequencing.